| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,684,349 – 7,684,422 |
| Length | 73 |
| Max. P | 0.888632 |

| Location | 7,684,349 – 7,684,422 |
|---|---|
| Length | 73 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.56674 |
| G+C content | 0.51807 |
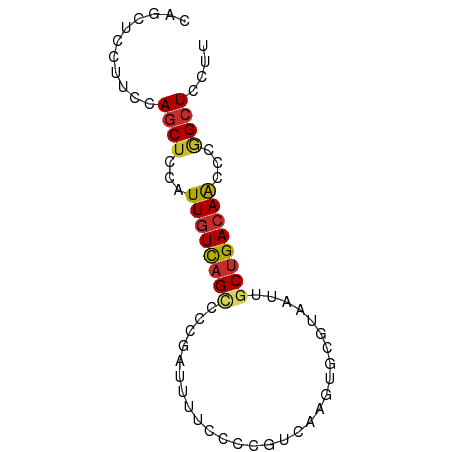
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -9.13 |
| Energy contribution | -9.27 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.18 |
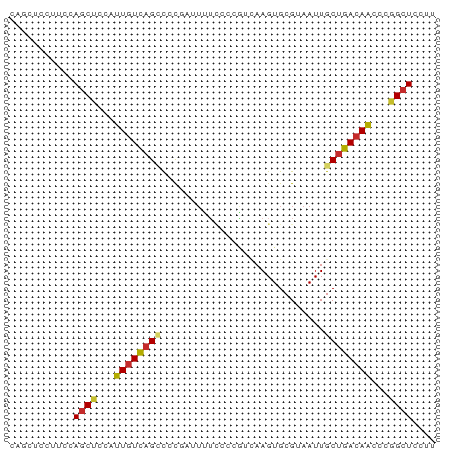
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
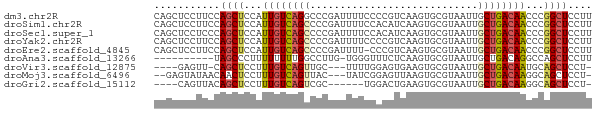
>dm3.chr2R 7684349 73 - 21146708 CAGCUCCUUCCAGCUCCAUUGUCAGGCCCGAUUUUCCCCGUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...(((((((...(((((.....(((.....))).))))))))))))...)))).... ( -12.70, z-score = 0.17, R) >droSim1.chr2R 6212124 73 - 19596830 CAGCUCCUUCCAGCUCCAUUGUCAGCCCCGAUUUUCCACAUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...((((((((...((((...(((.....)))...))))))))))))...)))).... ( -17.10, z-score = -2.76, R) >droSec1.super_1 5236377 73 - 14215200 CAGCUCCUCCCAGCUCCAUUGUCAGCCCCGAUUUUCCACAUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...((((((((...((((...(((.....)))...))))))))))))...)))).... ( -17.10, z-score = -2.80, R) >droYak2.chr2R 11869920 73 + 21139217 CAGCUCCUUCCAGCUCCAUUGUCAGCCCCGAUUUUCCCCGUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...((((((((...((((.....(((.....))).))))))))))))...)))).... ( -14.20, z-score = -1.44, R) >droEre2.scaffold_4845 4477064 72 + 22589142 CAGCUCCUUCCAGCUCCAUUGUCAGCCCCGAUUUU-CCCGUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...((((((((...((((..-..(((.....))).))))))))))))...)))).... ( -14.90, z-score = -1.73, R) >droAna3.scaffold_13266 6663609 62 + 19884421 ----------UAGCCCUUUUUUUUGGCCUUG-UGGGUUUCUCAAGUGCGUAAUUGCUGACAGGCCAGCUCCUU ----------............(((((((.(-(.(((....(......).....))).)))))))))...... ( -14.40, z-score = 0.43, R) >droVir3.scaffold_12875 15417360 64 + 20611582 ----GAGUU-CAGCUCCUUUGUCAGUUGC---UUUUGGAGUGAAGUGCGUAAUUGCUGACAAUGCAGCUCCU- ----(((((-..((....(((((((((((---(((......)))))).......)))))))).)))))))..- ( -16.51, z-score = -0.26, R) >droMoj3.scaffold_6496 24899641 67 - 26866924 --GAGUAUAACAACUCCUUUGUCAGUUAC---UAUCGGAGUUAAGUGCGUAAUUGCUGACAAGGCAGCUCCU- --((((......))))((((((((((...---...((.(......).)).....))))))))))........- ( -16.10, z-score = -1.00, R) >droGri2.scaffold_15112 4962334 62 + 5172618 ----CAGUUACAGCUCCUUUGUCAGUCGC------UGGACUGAAGUGCGUAAUUGCUGACAAGGCAGCUCCU- ----.......((((.(((((((((((((------...........))).....)))))))))).))))...- ( -20.10, z-score = -1.22, R) >consensus CAGCUCCUUCCAGCUCCAUUGUCAGCCCCGAUUUUCCCCGUCAAGUGCGUAAUUGCUGACAACCCGGCUCCUU ...........((((...((((((((............................))))))))...)))).... ( -9.13 = -9.27 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:14 2011