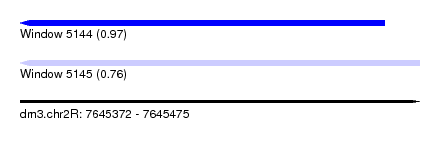
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,645,372 – 7,645,475 |
| Length | 103 |
| Max. P | 0.973585 |

| Location | 7,645,372 – 7,645,466 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Shannon entropy | 0.17793 |
| G+C content | 0.44746 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
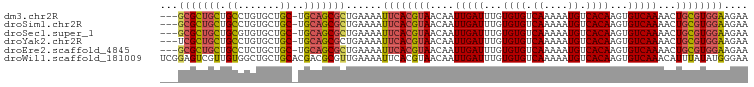
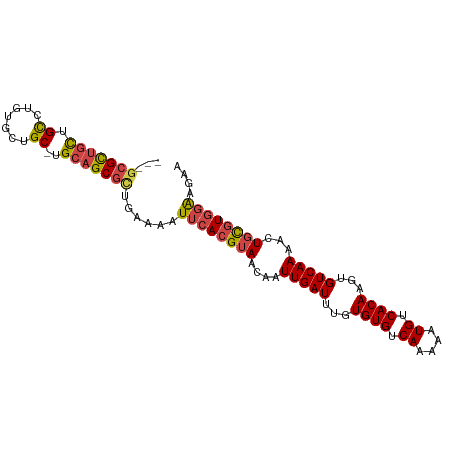
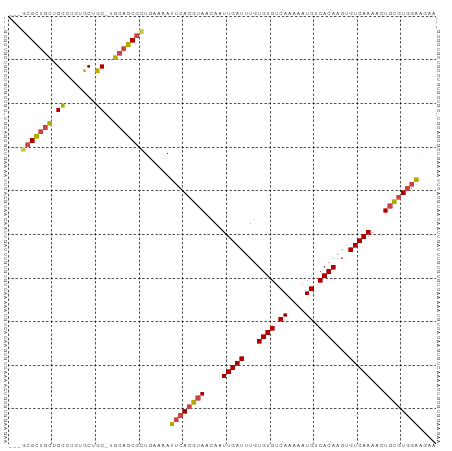
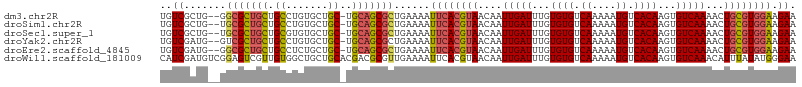
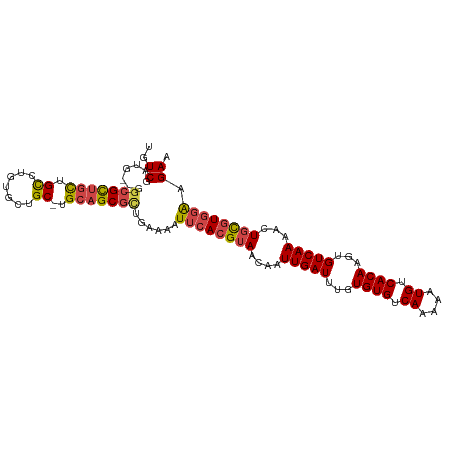
>dm3.chr2R 7645372 94 - 21146708 ---GCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ---(((((((.((.......))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... ( -34.50, z-score = -3.34, R) >droSim1.chr2R 6172497 94 - 19596830 ---GCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ---(((((((.((.......))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... ( -34.50, z-score = -3.34, R) >droSec1.super_1 5197648 94 - 14215200 ---GCGCUGCUGCGUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ---(((((((.(((.....)))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... ( -35.70, z-score = -3.20, R) >droYak2.chr2R 11830027 94 + 21139217 ---UCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ---.((((((.((.......))-.)))))).......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... ( -30.40, z-score = -2.42, R) >droEre2.scaffold_4845 4437935 94 + 22589142 ---GCGCUGCUGCCUCUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ---(((((((.((.......))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... ( -34.50, z-score = -3.68, R) >droWil1.scaffold_181009 3251889 98 + 3585778 UCGGAGUCGUUGUGGCUGCUGCACGACGCGUUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAACAUUUAUAUGGGAA ((((.(.(((((((.(....))))))))).))))...((((..(((...(((((...((((.((....)).))))...)))))....)))..)))).. ( -22.50, z-score = 0.34, R) >consensus ___GCGCUGCUGCCUGUGCUGC_UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ...(((((((.((....)).....)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).... (-26.12 = -26.65 + 0.53)
| Location | 7,645,372 – 7,645,475 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Shannon entropy | 0.18873 |
| G+C content | 0.45753 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.83 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7645372 103 - 21146708 UGUCGCUG--GGCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((..(.--((((((((.((.......))-.)))))))).)...((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). ( -36.40, z-score = -2.58, R) >droSim1.chr2R 6172497 103 - 19596830 UGUCGCUG--UGCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((....--.(((((((.((.......))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). ( -34.70, z-score = -2.31, R) >droSec1.super_1 5197648 103 - 14215200 UGUCGCUG--UGCGCUGCUGCGUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((....--.(((((((.(((.....)))-.)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). ( -35.90, z-score = -2.15, R) >droYak2.chr2R 11830027 103 + 21139217 UGUCGAUG--GUCGCUGCUGCCUGUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((...(--(.((((((.((.......))-.)))))))).....((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). ( -32.20, z-score = -1.83, R) >droEre2.scaffold_4845 4437935 103 + 22589142 UGUCGAUG--GGCGCUGCUGCCUCUGCUGC-UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((..(.--((((((((.((.......))-.)))))))).)...((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). ( -36.60, z-score = -3.18, R) >droWil1.scaffold_181009 3251889 106 + 3585778 CAUCGAUGUCGGAGUCGUUGUGGCUGCUGCACGACGCGUUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAACAUUUAUAUGGGAA ..(((((((....(((((.(..(...)..)))))))))))))...((((..(((...(((((...((((.((....)).))))...)))))....)))..)))).. ( -24.90, z-score = 0.52, R) >consensus UGUCGAUG__GGCGCUGCUGCCUGUGCUGC_UGCAGCGCUGAAAAUUCACGUAACAAUUGAUUUGUGUGUCAAAAAUGUCACAAGUGUCAAAACUGCGUGGAAGAA ..((.......(((((((.((....)).....)))))))......((((((((....(((((...((((.((....)).))))...)))))...)))))))).)). (-26.30 = -26.83 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:05 2011