
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,634,697 – 7,634,794 |
| Length | 97 |
| Max. P | 0.827957 |

| Location | 7,634,697 – 7,634,794 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.04 |
| Shannon entropy | 0.76224 |
| G+C content | 0.52145 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -10.28 |
| Energy contribution | -9.24 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
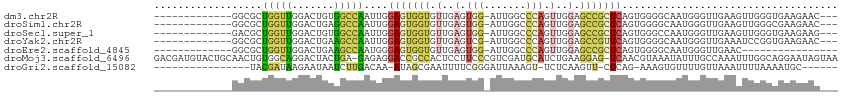
>dm3.chr2R 7634697 97 + 21146708 -------------GGCGCUGGUUGGACUGUGGCCAAUUGGAGUGGUGUUGAGUGG-AUUGGCCCAGUUGGAGCCGCUCAGUGGGGCAAUGGGUUGAAGUUGGGUGAAGAAC--- -------------.((.(.(.((.((((...(((.....(((((((.(..(.(((-......))).)..).))))))).....)))....)))).)).).).)).......--- ( -32.40, z-score = -1.66, R) >droSim1.chr2R 6161028 97 + 19596830 -------------GGCGCUGGUUGGACUGAGGCCAAUUGGAGUGGUGUUGAGUGG-AUUGGCCCAGUUGGAGCCGCUCAGUGGGGCAAUGGGUUGAAGUUGGGCGAAGAAC--- -------------.((.(.(.((.((((.(.(((.....(((((((.(..(.(((-......))).)..).))))))).....)))..).)))).)).).).)).......--- ( -34.60, z-score = -2.15, R) >droSec1.super_1 5186658 97 + 14215200 -------------GACGCUGGUUGGACUGUGGCCAAUUGGAGUGGUGUUGAGUGG-AUUGGCCCAGUUGGAGCCGCUCAGUGGGCCAAUGGGUUGAAGUUGGGUGAAGAAG--- -------------..(((((.((.((((.(((((.....(((((((.(..(.(((-......))).)..).)))))))....)))))...)))).)).)..))))......--- ( -36.80, z-score = -2.98, R) >droYak2.chr2R 11819104 97 - 21139217 -------------GGCGCUGGUUGGACUGAAGCCAAUUGGAGUGGUGUUGAGUCG-AUUGGCCCAGUUGGAGCCGUUCAGUGGGGCAAUGGGUUGAAAUCCGGUGAAGAAC--- -------------(((.((..((((......(((((((((..(......)..)))-))))))))))..)).))).((((.((((.(((....)))...)))).))))....--- ( -32.70, z-score = -1.47, R) >droEre2.scaffold_4845 4427020 84 - 22589142 -------------GGCGCUGGUUGGACUGAAGCCAAUGGGAGUGGUGUUGAGUGG-AUUGGCCCAGUUGGAGCCGCUCAGUGGGGCAAUGGGUUGAAC---------------- -------------.......(((.((((.(.(((.....(((((((.(..(.(((-......))).)..).))))))).....)))..).)))).)))---------------- ( -30.00, z-score = -1.60, R) >droMoj3.scaffold_6496 24829985 112 + 26866924 GACGAUGUACUGCAACUGUGGCAGGACUACUGA-GAGAGGACCGCCACUCCUUCCCGUCGAUGCAUCUGAAGGAG-UCAACGUAAAUAUUUGCCAAAUUUGGCAGGAAUAGUAA .(((.((.((.......(((((.(..((.....-...))..).)))))((((((..((......))..)))))))-))).))).....(((((((....)))))))........ ( -28.10, z-score = -0.10, R) >droGri2.scaffold_15082 7444 88 - 42191 ----------------UACGAUAAGAAUAAUCUUGACAA-AUAGCGAAUUUUCGGGAUUAAAGU-UCUCAAGUU-CUCAG-AAAGUGUUUUGUUAAAUUUUAAAAUGC------ ----------------.......(((((((((((((.((-((.....))))))))))))...))-)))......-.....-...((((((((........))))))))------ ( -11.20, z-score = 0.60, R) >consensus _____________GGCGCUGGUUGGACUGAGGCCAAUUGGAGUGGUGUUGAGUGG_AUUGGCCCAGUUGGAGCCGCUCAGUGGGGCAAUGGGUUGAAGUUGGGUGAAGAAC___ ..................(((((.......))))).....................((((.((((.((((......)))))))).))))......................... (-10.28 = -9.24 + -1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:04 2011