| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,619,427 – 7,619,518 |
| Length | 91 |
| Max. P | 0.977542 |

| Location | 7,619,427 – 7,619,518 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.42 |
| Shannon entropy | 0.66714 |
| G+C content | 0.45114 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -8.49 |
| Energy contribution | -8.28 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
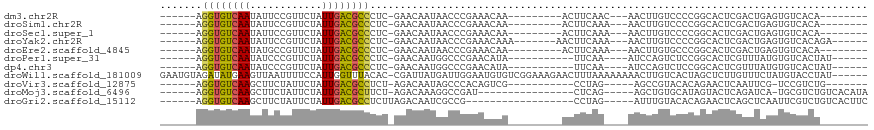
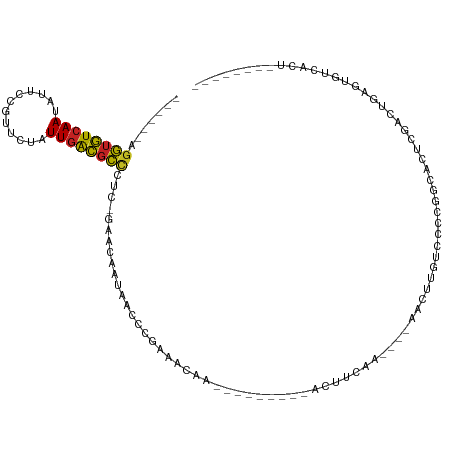
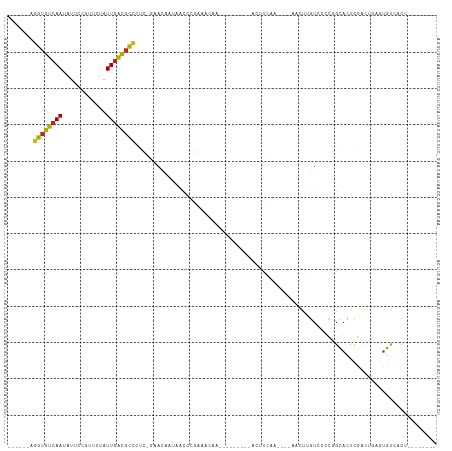
>dm3.chr2R 7619427 91 + 21146708 ------AGGUGUCAAUAUUCCGUUCUAUUGACGCCCUC-GAACAAUAACCCGAAACAA---------ACUUCAAC---AACUUGUCCCCGGCACUCGACUGAGUGUCACA-------- ------.((((((((((........))))))))))...-............(((....---------..)))...---...........(((((((....)))))))...-------- ( -23.60, z-score = -3.04, R) >droSim1.chr2R 6145247 91 + 19596830 ------AGGUGUCAAUAUUCCGUUCUAUUGACGCCCUC-GAACAAUAACCCGAAACAA---------ACUUCAAA---AACUUGUCCCCGGCACUCGACUGAGUGUCACA-------- ------.((((((((((........))))))))))...-............(((....---------..)))...---...........(((((((....)))))))...-------- ( -23.60, z-score = -3.24, R) >droSec1.super_1 5171331 91 + 14215200 ------AGGUGUCAAUAUUCCGUUCUAUUGACGCCCUC-GAACAAUAACCCGAAACAA---------ACUUCAAA---AACUUGUCCCCGGCACUCGACUGAGUGUCACA-------- ------.((((((((((........))))))))))...-............(((....---------..)))...---...........(((((((....)))))))...-------- ( -23.60, z-score = -3.24, R) >droYak2.chr2R 11803624 95 - 21139217 ------AGGUGUCAAUAUUCCGUUCUAUUGACGCCCUC-GAACAAUAACCCGAAACAAA-------AACUUCAAA---AACUUGUCCCCGGCACUCGACUGAGUGUCACAGA------ ------.((((((((((........)))))))))).((-(..........)))......-------.........---...........(((((((....))))))).....------ ( -23.40, z-score = -2.76, R) >droEre2.scaffold_4845 4411074 91 - 22589142 ------AGGUGUCAAUAUGCCGUUCUAUUGACGCCCUC-GAACAAUAACCCGAAACAA---------ACUUCAAA---AACUUGUGCCCGGCACUCGACUGAGUGUCACA-------- ------.((((((((((........))))))))))...-..((((......(((....---------..)))...---...))))....(((((((....)))))))...-------- ( -23.90, z-score = -2.66, R) >droPer1.super_31 79844 90 - 935084 ------AGGUGUCAAUAUCCCGUUCUAUUGACGCCCUC-GAACAAUGGCCCGAACAUA-----------UUCAA----AUCCAGUCUCCGGCACUCGUUUAUGUGUCACUAU------ ------.((((((((((........))))))))))...-((....(((...(((....-----------)))..----..))).))...(((((........))))).....------ ( -21.50, z-score = -2.12, R) >dp4.chr3 16896991 90 + 19779522 ------AGGUGUCAAUAUCCCGUUCUAUUGACGCCCUC-GAACAAUGGCCCGAACAUA-----------UUCAA----AUCCAGUCUCCGGCACUCGUUUAUGUGUCACUAU------ ------.((((((((((........))))))))))...-((....(((...(((....-----------)))..----..))).))...(((((........))))).....------ ( -21.50, z-score = -2.12, R) >droWil1.scaffold_181009 3224388 111 - 3585778 GAAUGUAGAUAUGAAGUUAAUUUUCCAUUGGUUUACAC-CGAUUAUGAUUGGAAUGUGUCGGAAAGAACUUUAAAAAAAACUUGUCACUAGCUCUUGUUUCUAUGUACCUAU------ ..(((((((..(((((((...(((((((((((....))-))))...(((........)))))))).)))))))................(((....))))))))))......------ ( -19.10, z-score = -0.33, R) >droVir3.scaffold_12875 15326097 87 - 20611582 ------AGGUGUCAAGCUUCUAUUCUAUUGACGCCUCU-AGACAAUAGCCCACAGUCG-----------CCUAG-----AGCCGUACACAGAACUCAAUUCG-UCCGUCUG------- ------(((((((((............))))))))).(-((((....((........)-----------)...(-----((............)))......-...)))))------- ( -16.10, z-score = -0.12, R) >droMoj3.scaffold_6496 24808922 89 + 26866924 ------AGGUGUCAAGCUUCUAUUCUAUUGACGCUUCU-AGACAAAGGCCGAU----------------CUCAG-----AGCUGUGCAUAGUACUCAGAUCA-UGCGUCUGUCACAUA ------(((((((((............)))))))))..-.((((.(.((.(((----------------((.((-----.((((....)))).)).))))).-.)).).))))..... ( -24.90, z-score = -1.09, R) >droGri2.scaffold_15112 4880602 89 - 5172618 ------AGGUGUCAAGCUUCUAUUCUAUUGACGCCUCUUAGACAAUCGCCG------------------CCUAG-----AUUUGUACACAGAACUCAGCUCAAUUCGUCUGUCACUUC ------(((((((((............)))))))))....((((..((..(------------------(..((-----.((((....)))).))..))......))..))))..... ( -19.70, z-score = -1.58, R) >consensus ______AGGUGUCAAUAUUCCGUUCUAUUGACGCCCUC_GAACAAUAACCCGAAACAA_________ACUUCAA____AACUUGUCCCCGGCACUCGACUGAGUGUCACU________ .......((((((((............))))))))................................................................................... ( -8.49 = -8.28 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:16:00 2011