| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,594,543 – 2,594,680 |
| Length | 137 |
| Max. P | 0.606378 |

| Location | 2,594,543 – 2,594,680 |
|---|---|
| Length | 137 |
| Sequences | 11 |
| Columns | 161 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Shannon entropy | 0.36522 |
| G+C content | 0.37303 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
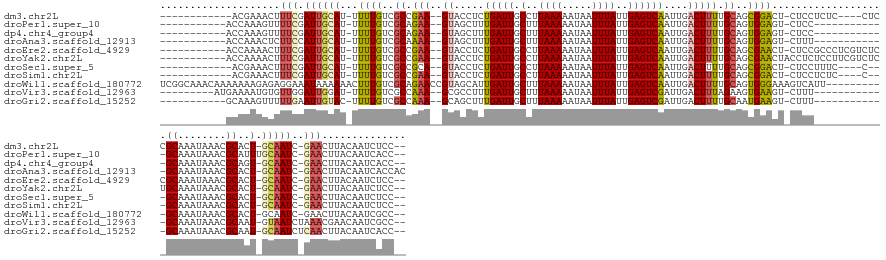
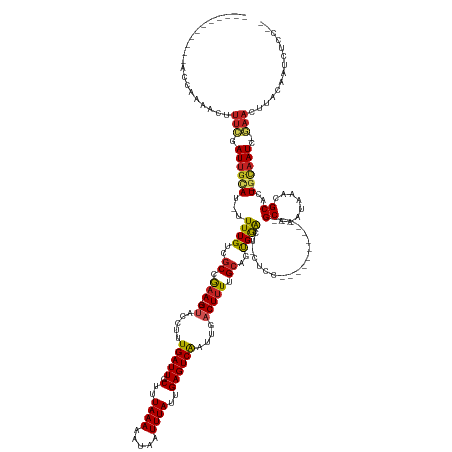
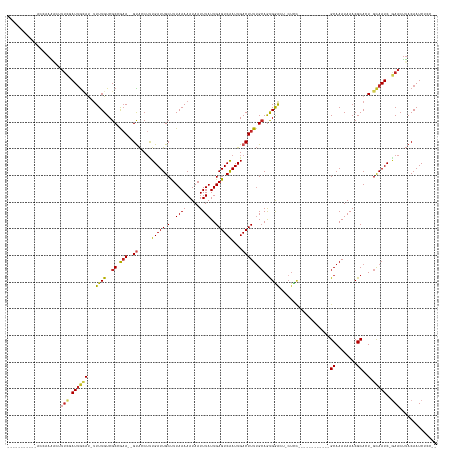
>dm3.chr2L 2594543 137 + 23011544 ------------ACGAAACUUUCGAUUGCAU-UUUUGUCGCCGAA--GUACCUCUGAUUGCCUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGCGGACU-CUCCUCUC----CUCCGCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCUCC-- ------------........((((((((((.-.......((.(((--((......(((((.(((((.((....)).))))).))))).))))).)).(((((..-........----.)))))............)-))))))-)))............-- ( -31.30, z-score = -3.60, R) >droPer1.super_10 907006 131 + 3432795 -----------ACCAAAGUUUUCGAUUGCAU-UUUUGUCGCAGAA--GUAGCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGUGGAGU-CUCC------------GCAAAUAAACGCAUUUGCAAUC-GAACUUACAAUCACC-- -----------....((((..(((((((((.-...(((.((((((--((....((((((.(..((((.....))))..)))))))...)))))))).((((...-..))------------))........)))..)))))))-)))))).........-- ( -37.20, z-score = -3.90, R) >dp4.chr4_group4 1899777 130 + 6586962 -----------ACCAAAGUUUUCGAUUGCAU-UUUUGUCGCAGAA--GUAGCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGUGGAGU-CUCC------------GCAAAUAAACGCAGU-GCAAUC-GAACUUACAAUCACC-- -----------....((((..((((((((((-((((((.((((((--((....((((((.(..((((.....))))..)))))))...)))))))).((((...-..))------------))..)))))...)))-))))))-)))))).........-- ( -37.60, z-score = -3.84, R) >droAna3.scaffold_12913 405945 132 - 441482 -----------ACCAAACUCUUCCAUUGCAU-UUUUGUCGCAAAA--GUAGCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGUGGAGU-CUUU------------GCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCACCAC -----------.........(((.((((((.-(((..(.((((((--((....((((((.(..((((.....))))..)))))))...)))))))).)..))).-...(------------((........))).)-))))).-))).............. ( -26.60, z-score = -1.28, R) >droEre2.scaffold_4929 2639128 142 + 26641161 -----------ACCAAAACUUUCGAUUGCAU-UUUUGUCGCCGAA--GUACCUCUGAUUGCCUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGCGAACU-CUCCGCCCUCGUCUCCGCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCUCC-- -----------.........((((((((((.-.(((((.((.(((--((......(((((.(((((.((....)).))))).))))).))))).)).)))))..-................((........))..)-))))))-)))............-- ( -28.80, z-score = -2.94, R) >droYak2.chr2L 2582534 143 + 22324452 -----------ACCAAAACUUUCGAUUGCAU-UUUUGUCGCCGAA--GUACCUCUGAUUGCCUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGCGAACUACCUCUCCUUCGUCUCUGCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCUCC-- -----------.........((((((((((.-.(((((.((.(((--((......(((((.(((((.((....)).))))).))))).))))).)).)))))..................(((........))).)-))))))-)))............-- ( -29.00, z-score = -3.44, R) >droSec1.super_5 762856 134 + 5866729 ------------ACGAAACUUUCGAUUGCAU-UUUUGUCGCCGCA--GUACCUCUGAUUGCCUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGCGGACU-CUCCUUUC----C---GCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCUCC-- ------------........(((((((((((-(((((..((..((--(.....)))...))..)))))))..(((((((((((....))))).....(((((..-......))----)---)).))))))......-))))))-)))............-- ( -33.60, z-score = -4.03, R) >droSim1.chr2L 2552919 134 + 22036055 ------------ACGAAACUUUCGAUUGCAU-UUUUGUCGCCGAA--GUACCUCUGAUUGCCUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGCGGACU-CUCCUCUC----C---GCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCUCC-- ------------........((((((((((.-.......((.(((--((......(((((.(((((.((....)).))))).))))).))))).)).(((((..-......))----)---))............)-))))))-)))............-- ( -32.30, z-score = -3.79, R) >droWil1.scaffold_180772 1238313 147 + 8906247 UCGGCAAACAAAAAAAGAGAGGAAAUAAAAAACUUUGUCGCAGAACCGUAGCAUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGUGGGAAAGUCAUU----------GCAAAUAAACGCACU-GCAAUC-GAACUUACAAUCGCC-- ..(((...........(.((.(((.........))).)).)......((((..((((((((.................(((((....)))))(((((((((.....)))))----------))))...........-))))))-))..))))....)))-- ( -31.00, z-score = -1.09, R) >droVir3.scaffold_12963 11166875 133 + 20206255 ---------AUGAAAAUGUGUUGGAUUGGAU-UUUUGUCGCCAAA--GCGCCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCGAUUGACUUUAGAAGUGAAGU-CUUU------------GCAAAUAAACGCAAU-GUAAUCUAAACGAACAAUCGCC-- ---------.........((((.(.((((((-(.(((.((..(((--(((........))))))........((((((..(.(((..(((((((....))))))-).))------------)).))))))))))).-..))))))).).))))......-- ( -24.80, z-score = -0.01, R) >droGri2.scaffold_15252 5301638 131 + 17193109 -----------GCAAAGUUUUUGAAUUGUAC-UUUUGUCGCCAAA--GCAGCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCGAUUGACUUUUGCAAUGAAGU-CUUU------------GCAAAUAAACGCAAU-GCAAUCUCAACUUACAAUCACC-- -----------.....((..((((((((((.-..(((.((..(((--((((......)))))))........(((((((((((....))))).(((((......-..))------------)))))))))))))))-))))).))))...)).......-- ( -22.60, z-score = 0.40, R) >consensus ___________ACCAAAACUUUCGAUUGCAU_UUUUGUCGCCGAA__GUACCUUUGAUUGCUUUAAAAAUAAUUUAUUGAGUCAAUUGACUUUUGCAGUGGACU_CUCC____________GCAAAUAAACGCACU_GCAAUC_GAACUUACAAUCUCC__ ...............................................(((..((.((((((.................(((((....))))).............................((........))....)))))).))...)))......... (-12.25 = -12.35 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:46 2011