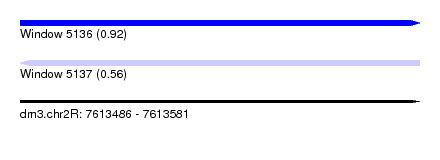
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,613,486 – 7,613,581 |
| Length | 95 |
| Max. P | 0.922301 |

| Location | 7,613,486 – 7,613,581 |
|---|---|
| Length | 95 |
| Sequences | 6 |
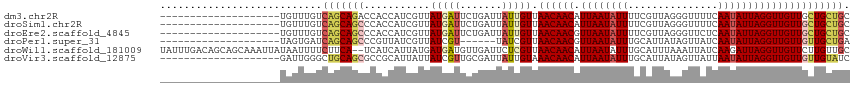
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.46562 |
| G+C content | 0.33552 |
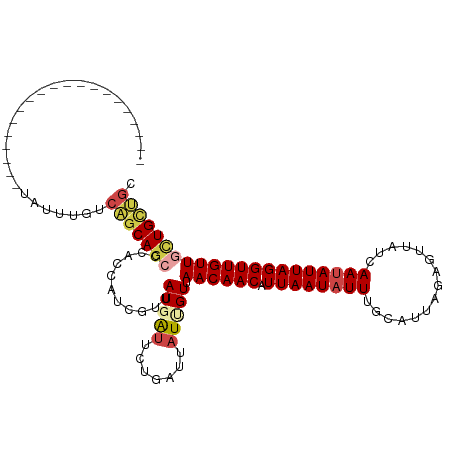
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -8.25 |
| Energy contribution | -9.78 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
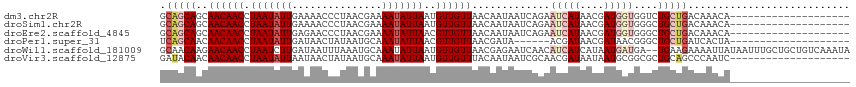
>dm3.chr2R 7613486 95 + 21146708 GCAGCAGCAACAACCUAAUAUUGAAAACCCUAACGAAAAUAUUAAUGUUGUUAACAAUAAUCAGAAUCAUAACGAUGGUGGUCUGCUGACAAACA-------------------- .((((((.((((((.(((((((...............)))))))..)))))).......((((..(((.....)))..)))))))))).......-------------------- ( -19.16, z-score = -2.32, R) >droSim1.chr2R 6137465 95 + 19596830 GCAGCAGCAACAACCUAAUAUUGAAAACCCUAACGAAAAUAUUAAUGUUGUUAACAAUAAUCAGAAUCAUAACGAUGGUGGGCUGCUGACAAACA-------------------- .(((((((((((((.(((((((...............)))))))..))))))........(((..(((.....)))..)))))))))).......-------------------- ( -23.06, z-score = -3.51, R) >droEre2.scaffold_4845 4405494 95 - 22589142 GCAGCAGCAACAACCUAAUAUUGAGAACCCUAACGAAAAUAUUAACGUUGUUAACAAUAAUCAGAAUCAUAACGAUGGUGGGCUGCUGACAAACA-------------------- .(((((((((((((.(((((((..(........)...)))))))..))))))........(((..(((.....)))..)))))))))).......-------------------- ( -23.30, z-score = -3.53, R) >droPer1.super_31 73031 89 - 935084 UCAGCAACAACAACCUAAUAUUGAUAACUAUAAUGCAAAUAUUAACGUUGUUAACGAUA------ACGAUAACGAUAACGGGCUGCUGAUCACUA-------------------- ((((((.(((((((.(((((((...............)))))))..))))))..((.((------.((....)).)).)).).))))))......-------------------- ( -16.26, z-score = -2.64, R) >droWil1.scaffold_181009 3213495 113 - 3585778 GCAACAAGAACAACCUAAUCUUGAUAAUUUAAAUGCAAAUAUUAAUGUUGUUAACGAGAAUCAACAUCAUCAUAAUGAUGA--UGAAGAAAAUUAUAAUUUGCUGCUGUCAAAUA (((.(((((.........)))))...........((((((..((((.(((....)))...((..(((((((.....)))))--))..))..))))..)))))))))......... ( -21.00, z-score = -1.97, R) >droVir3.scaffold_12875 15317358 95 - 20611582 GAUACAACAACAACCUAAUAUUAAUAACUAUAAUGCAAAUAUUAAUGUUGUUUACAAUAAUCGCAACGAUAAUAAUGCGGCGCUGCAGCCCAAUC-------------------- ..................................(((..(((((.((((((...........)))))).))))).)))(((......))).....-------------------- ( -13.40, z-score = -0.42, R) >consensus GCAGCAACAACAACCUAAUAUUGAUAACCCUAACGAAAAUAUUAAUGUUGUUAACAAUAAUCAGAAUCAUAACGAUGGUGGGCUGCUGACAAACA____________________ .(((((..((((((.(((((((...............)))))))..)))))).............(((((....)))))....)))))........................... ( -8.25 = -9.78 + 1.53)
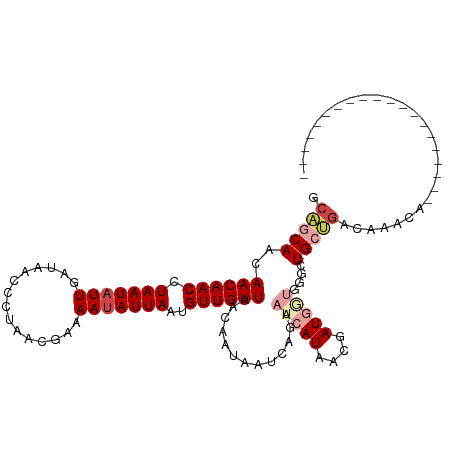
| Location | 7,613,486 – 7,613,581 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.46562 |
| G+C content | 0.33552 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -11.58 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7613486 95 - 21146708 --------------------UGUUUGUCAGCAGACCACCAUCGUUAUGAUUCUGAUUAUUGUUAACAACAUUAAUAUUUUCGUUAGGGUUUUCAAUAUUAGGUUGUUGCUGCUGC --------------------.......((((((((.((((..(((..(((((((((...((((((.....)))))).....)))))))))...)))..).))).))..)))))). ( -18.00, z-score = -0.33, R) >droSim1.chr2R 6137465 95 - 19596830 --------------------UGUUUGUCAGCAGCCCACCAUCGUUAUGAUUCUGAUUAUUGUUAACAACAUUAAUAUUUUCGUUAGGGUUUUCAAUAUUAGGUUGUUGCUGCUGC --------------------.......(((((((..((((..(((..(((((((((...((((((.....)))))).....)))))))))...)))..).)))....))))))). ( -19.90, z-score = -0.99, R) >droEre2.scaffold_4845 4405494 95 + 22589142 --------------------UGUUUGUCAGCAGCCCACCAUCGUUAUGAUUCUGAUUAUUGUUAACAACGUUAAUAUUUUCGUUAGGGUUCUCAAUAUUAGGUUGUUGCUGCUGC --------------------.......(((((((..((.(((..((((((((((((...(((((((...))))))).....)))))))))....)))...))).)).))))))). ( -21.80, z-score = -1.51, R) >droPer1.super_31 73031 89 + 935084 --------------------UAGUGAUCAGCAGCCCGUUAUCGUUAUCGU------UAUCGUUAACAACGUUAAUAUUUGCAUUAUAGUUAUCAAUAUUAGGUUGUUGUUGCUGA --------------------(((..(.((((((((((((...((((.((.------...)).)))))))).(((((((.((......))....))))))))))))))))..))). ( -21.90, z-score = -2.79, R) >droWil1.scaffold_181009 3213495 113 + 3585778 UAUUUGACAGCAGCAAAUUAUAAUUUUCUUCA--UCAUCAUUAUGAUGAUGUUGAUUCUCGUUAACAACAUUAAUAUUUGCAUUUAAAUUAUCAAGAUUAGGUUGUUCUUGUUGC .........(((((((..........((..((--(((((.....)))))))..))........((((((.(((((.((((.((.......))))))))))))))))).))))))) ( -23.10, z-score = -1.78, R) >droVir3.scaffold_12875 15317358 95 + 20611582 --------------------GAUUGGGCUGCAGCGCCGCAUUAUUAUCGUUGCGAUUAUUGUAAACAACAUUAAUAUUUGCAUUAUAGUUAUUAAUAUUAGGUUGUUGUUGUAUC --------------------........((((((..((((..........)))).........((((((.((((((((.((......))....)))))))))))))))))))).. ( -19.50, z-score = -0.12, R) >consensus ____________________UAUUUGUCAGCAGCCCACCAUCGUUAUGAUUCUGAUUAUUGUUAACAACAUUAAUAUUUGCAUUAGAGUUAUCAAUAUUAGGUUGUUGCUGCUGC ...........................(((((((...........(((((.......))))).((((((.((((((((...............))))))))))))))))))))). (-11.58 = -12.44 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:59 2011