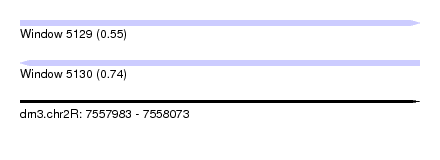
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,557,983 – 7,558,073 |
| Length | 90 |
| Max. P | 0.742735 |

| Location | 7,557,983 – 7,558,073 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.45319 |
| G+C content | 0.35988 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -9.48 |
| Energy contribution | -9.15 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
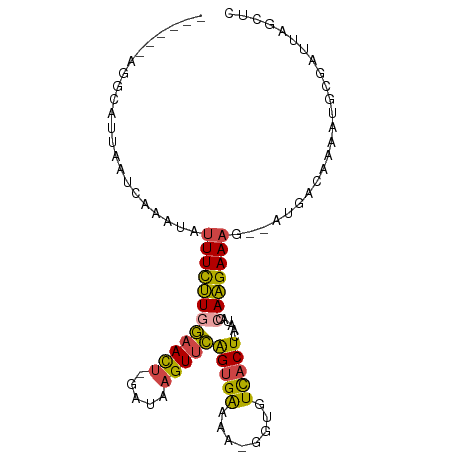
>dm3.chr2R 7557983 90 + 21146708 ------AGGCAUUAAUCAAAUAUUUCUUAGAACU-GAUAAGUUCAGCGAAAAAGGUGUCACUCAAUACCAAGAAAG--AUGACAAAAUGCGAUUAGCUC ------..(((((..(((....((((((.(((((-....))))).........(((((......))))))))))).--.)))...)))))......... ( -19.70, z-score = -2.44, R) >droSim1.chr2R 6080257 89 + 19596830 ------AGGCAUUAAUCAAAUAUUUCUUGGAACU-GAUAAGUUCAGUGGAAA-GGUGUCACUCAAUACCAAGAAAG--AUGACAAAAUGCGAUUAGCUC ------..(((((..(((....((((((((((((-....)))))(((((...-....)))))......))))))).--.)))...)))))......... ( -20.30, z-score = -1.92, R) >droSec1.super_1 5109553 89 + 14215200 ------GGGCAUUAAUCAAAUAUUUCUUGGAACU-GAUAAGUUCAGUGAAAA-GGUGUCACUCAAUACCAAGAAAG--AUGACAAAAUGCGAUUAGCUC ------..(((((..(((....((((((((((((-....)))))(((((...-....)))))......))))))).--.)))...)))))......... ( -21.20, z-score = -2.43, R) >droYak2.chr2R 11742369 89 - 21139217 ------UAUCGUUAAUCAAACAGUUCUUGGAACU-GAUAAGUUCAGUGAAAG-GCUGUCACUCAAUACCAAGAAAG--AUCACGAAACGCGAUUGGCUU ------....(((((((......(((((((((((-....)))))(((((...-....)))))......))))))..--.....(.....)))))))).. ( -19.10, z-score = -1.02, R) >droEre2.scaffold_4845 4352841 87 - 22589142 ------UGUCAUUAAUC--ACCUUUCUUGGAACA-GAUAAGUUCGGUGAAGG-GGUGUCACUCGAUACCAAGAAAG--AUUACAAAAGGCGAUUGGCUU ------......(((((--.(((((..(.((((.-.....)))).).)))))-((((((....))))))......)--))))....((((.....)))) ( -22.20, z-score = -1.44, R) >triCas2.ChLG8 9450362 97 + 15773733 ACAGAAAAUCGCUAAUUAAAUAUUUUGUAUUAUUUGUUGAGUUGGGUGACUG-AACUUUACUCAAUGACAUGAAAACAAUAAAAAUACACCGUUGACU- .........................((((((.(((((((.(((((((((...-....)))))))))....(....)))))))))))))).........- ( -14.00, z-score = -0.36, R) >consensus ______AGGCAUUAAUCAAAUAUUUCUUGGAACU_GAUAAGUUCAGUGAAAA_GGUGUCACUCAAUACCAAGAAAG__AUGACAAAAUGCGAUUAGCUC ......................((((((((((((.....)))))(((((........)))))......)))))))........................ ( -9.48 = -9.15 + -0.33)

| Location | 7,557,983 – 7,558,073 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.45319 |
| G+C content | 0.35988 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -9.26 |
| Energy contribution | -9.51 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7557983 90 - 21146708 GAGCUAAUCGCAUUUUGUCAU--CUUUCUUGGUAUUGAGUGACACCUUUUUCGCUGAACUUAUC-AGUUCUAAGAAAUAUUUGAUUAAUGCCU------ .........(((((..((((.--.((((((((.(((((((((........))))........))-))).))))))))....)))).)))))..------ ( -19.50, z-score = -2.03, R) >droSim1.chr2R 6080257 89 - 19596830 GAGCUAAUCGCAUUUUGUCAU--CUUUCUUGGUAUUGAGUGACACC-UUUCCACUGAACUUAUC-AGUUCCAAGAAAUAUUUGAUUAAUGCCU------ .........(((((..((((.--.((((((((.((((((((.....-....)))........))-))).))))))))....)))).)))))..------ ( -20.30, z-score = -2.64, R) >droSec1.super_1 5109553 89 - 14215200 GAGCUAAUCGCAUUUUGUCAU--CUUUCUUGGUAUUGAGUGACACC-UUUUCACUGAACUUAUC-AGUUCCAAGAAAUAUUUGAUUAAUGCCC------ .........(((((..((((.--.((((((((.(((((((((....-...))))........))-))).))))))))....)))).)))))..------ ( -21.90, z-score = -3.25, R) >droYak2.chr2R 11742369 89 + 21139217 AAGCCAAUCGCGUUUCGUGAU--CUUUCUUGGUAUUGAGUGACAGC-CUUUCACUGAACUUAUC-AGUUCCAAGAACUGUUUGAUUAACGAUA------ ..((.....))...(((((((--(.(((((((.(((((....(((.-......)))......))-))).)))))))......)))).))))..------ ( -18.60, z-score = -0.75, R) >droEre2.scaffold_4845 4352841 87 + 22589142 AAGCCAAUCGCCUUUUGUAAU--CUUUCUUGGUAUCGAGUGACACC-CCUUCACCGAACUUAUC-UGUUCCAAGAAAGGU--GAUUAAUGACA------ .....(((((((((((.....--.......(((.(((.((((....-...))))))))))....-........)))))))--)))).......------ ( -20.95, z-score = -2.41, R) >triCas2.ChLG8 9450362 97 - 15773733 -AGUCAACGGUGUAUUUUUAUUGUUUUCAUGUCAUUGAGUAAAGUU-CAGUCACCCAACUCAACAAAUAAUACAAAAUAUUUAAUUAGCGAUUUUCUGU -.((.......((((((((((((((((((......))).....(((-.(((......))).))))))))))).))))))).......)).......... ( -10.34, z-score = 0.46, R) >consensus GAGCCAAUCGCAUUUUGUCAU__CUUUCUUGGUAUUGAGUGACACC_CUUUCACUGAACUUAUC_AGUUCCAAGAAAUAUUUGAUUAAUGACU______ ..........((((..((((....((((((((...(((((.................))))).......))))))))....)))).))))......... ( -9.26 = -9.51 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:53 2011