| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,548,027 – 7,548,187 |
| Length | 160 |
| Max. P | 0.960337 |

| Location | 7,548,027 – 7,548,187 |
|---|---|
| Length | 160 |
| Sequences | 10 |
| Columns | 170 |
| Reading direction | forward |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.67718 |
| G+C content | 0.48282 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -24.14 |
| Energy contribution | -22.23 |
| Covariance contribution | -1.91 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
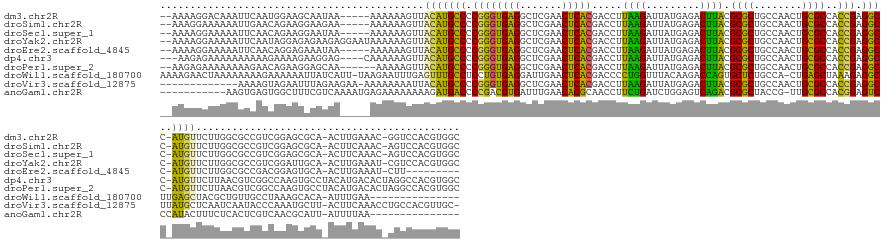
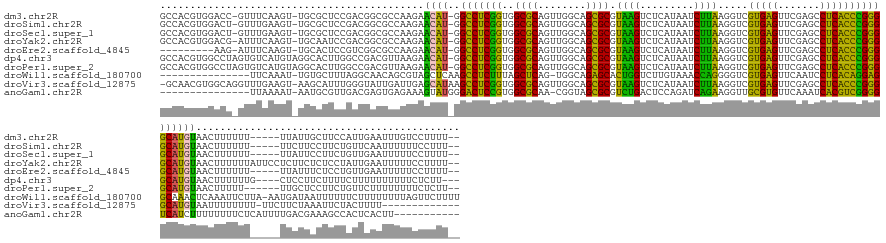
>dm3.chr2R 7548027 160 + 21146708 --AAAAGGACAAAUUCAAUGGAAGCAAUAA-----AAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUGGCGCCGUCGGAGCGCA-ACUUGAAAC-GGUCCACGUGGC --....((((...(((((............-----....(((((.((((...(...(((((.(((......)))))))).)..)))).)))))..(((((.((.((.((((((..(((...-...))).)))))).)).))))))).-..)))))..-.))))....... ( -48.00, z-score = -0.54, R) >droSim1.chr2R 6070625 160 + 19596830 --AAAGGAAAAAAUUGAACAGAAGGAAGAA-----AAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUGGCGCCGUCGGAGCGCA-ACUUCAAAC-AGUCCACGUGGC --...(((.....(((((............-----....(((((.((((...(...(((((.(((......)))))))).)..)))).)))))..(((((.((.((.((((((..(((...-...))).)))))).)).))))))).-..)))))..-..)))....... ( -44.10, z-score = 0.05, R) >droSec1.super_1 5099902 160 + 14215200 --AAAAGGAAAAAUUCAACAGAAGGAAUAA-----AAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUGGCGCCGUCGGAGCGCA-ACUUCAAAC-AGUCCACGUGGC --....(((...........((((......-----....(((((.((((...(...(((((.(((......)))))))).)..)))).)))))..(((((.((.((.((((((..(((...-...))).)))))).)).))))))).-.))))....-..)))....... ( -42.93, z-score = 0.06, R) >droYak2.chr2R 11732311 165 - 21139217 --AAAAGGAAAAAUUCAAUAGGAGAGAAGAGGAAUAAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUGGCGCCGUCGGAUUGCA-ACUUGAAAU-CGUCCACGUGGC --....(((....(((((...(..(...((((.......(((..(((((((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).)).)-)))).)))....)).))...)..).-..)))))..-..)))....... ( -39.40, z-score = 1.41, R) >droEre2.scaffold_4845 4343046 151 - 22589142 --AAAAGGAAAAAUUCAACAGGAGAAAUAA-----AAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUGGCGCCGACGGAGUGCA-ACUUGAAAU-CUU--------- --................((((........-----....(((((.((((...(...(((((.(((......)))))))).)..)))).)))))..(((((.((..(.((((((..(((...-...))).)))))).)..))))))).-.))))....-...--------- ( -37.70, z-score = -0.23, R) >dp4.chr3 4732446 162 + 19779522 ---AAGAGAAAAAAAAAAGAAAAGAAGGAG----CAAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUAACGUCGGCCAAGUGCCUACAUGACACUAGGCCACGUGGC ---..........................(----(....(((..(((((((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).)).)-)))).))).((((.((((.((((.(.....).)))).)))))))).)) ( -46.60, z-score = -1.51, R) >droPer1.super_2 4925227 161 + 9036312 --AAGAGAAAAAAAAGAACAGAAGGAGCAA------AAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC-AUGUUCUUAACGUCGGCCAAGUGCCUACAUGACACUAGGCCACGUGGC --...........(((((((...(.(((..------.....))).)..(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).-.))))))).((((.((((.((((.(.....).)))).))))))))... ( -48.10, z-score = -1.80, R) >droWil1.scaffold_180700 4631899 152 + 6630534 AAAAGAACUAAAAAAAAGAAAAAAUUAUCAUU-UAAGAAUUUGAGUUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCA-CUGAGCUAAAGAGGCUUGAGCUACGCUGUUGCCUAAAGCACA-AUUUGAA--------------- ...........................(((..-..........((((((((((.((((((.......))))))....((((((.....)))))).((((.....-..))))....)))))..)))))..(((.........)))...-...))).--------------- ( -33.50, z-score = -1.01, R) >droVir3.scaffold_12875 7716987 154 - 20611582 -------------AAAAGUAGAAUUUAGAAGAA-AAAAAAAAUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUUAUGCUCAAUCAAUACCCAAAUGCUU-ACUUCAAACCUGCCACGUUGC- -------------....((((..(((.((((..-..............(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).)))....((.................))..-.))))))).))))........- ( -30.53, z-score = -0.27, R) >anoGam1.chr2R 57350968 142 + 62725911 -----------AAGUGAGUGGCUUUCGUCAAAAUGAGAAAAAAAAGAUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCG-UUGCGCCACGGAGUCCCAUACUUUCUCACUCGUCAACGCAUU-AUUUUAA--------------- -----------..(((((((((....))).....((((((........(((.(((.((((.......))))......((((((.....)))))).((((.....-..))))..))).)))......))))))))))).)........-.......--------------- ( -32.94, z-score = 0.07, R) >consensus __AAAAGGAAAAAUUCAACAGAAGAAAUAA_____AAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCC_AUGUUCUUGGCGCCGUCGAAGCGCA_ACUUGAAAC_AGUCCACGUGGC ............................................(((((((.((((((((.......))))).....((((.........)))).((((........))))..))).)))..))))............................................ (-24.14 = -22.23 + -1.91)
| Location | 7,548,027 – 7,548,187 |
|---|---|
| Length | 160 |
| Sequences | 10 |
| Columns | 170 |
| Reading direction | reverse |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.67718 |
| G+C content | 0.48282 |
| Mean single sequence MFE | -47.03 |
| Consensus MFE | -27.92 |
| Energy contribution | -25.47 |
| Covariance contribution | -2.45 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
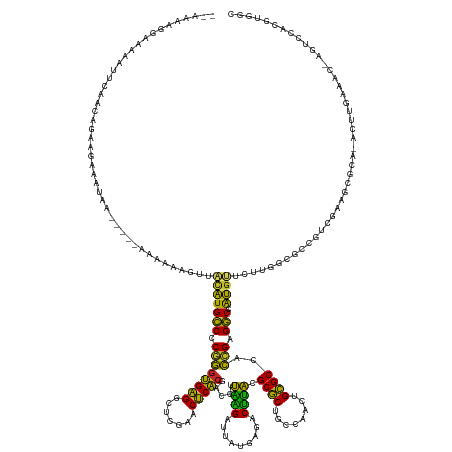
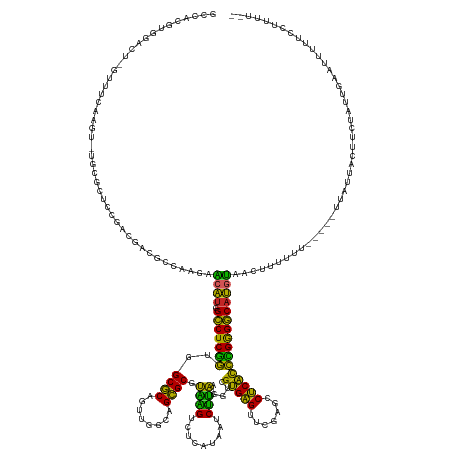
>dm3.chr2R 7548027 160 - 21146708 GCCACGUGGACC-GUUUCAAGU-UGCGCUCCGACGGCGCCAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUU-----UUAUUGCUUCCAUUGAAUUUGUCCUUUU-- .......((((.-..(((((..-.((((((((((.((((((.((....-....))..)))))).))))).)))))((((..((((..(((....)))..)))).....(((((....))))).............-----...)))).....)))))...))))....-- ( -55.90, z-score = -1.90, R) >droSim1.chr2R 6070625 160 - 19596830 GCCACGUGGACU-GUUUGAAGU-UGCGCUCCGACGGCGCCAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUU-----UUCUUCCUUCUGUUCAAUUUUUUCCUUU-- ......(((((.-(...((((.-.((((((((((.((((((.((....-....))..)))))).))))).)))))..((((((((..(((....)))..)))).....(((((....))))).....))))....-----..))))...).)))))............-- ( -52.30, z-score = -1.34, R) >droSec1.super_1 5099902 160 - 14215200 GCCACGUGGACU-GUUUGAAGU-UGCGCUCCGACGGCGCCAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUU-----UUAUUCCUUCUGUUGAAUUUUUCCUUUU-- (((.((.((...-(((((((..-.((((((((((.((((((.((....-....))..)))))).))))).)))))......((((..(((....)))..)))))))))))....)))).))).............-----............................-- ( -52.00, z-score = -1.13, R) >droYak2.chr2R 11732311 165 + 21139217 GCCACGUGGACG-AUUUCAAGU-UGCAAUCCGACGGCGCCAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUUAUUCCUCUUCUCUCCUAUUGAAUUUUUCCUUUU-- .......(((.(-(.(((((((-(((....((((.((((((.((....-....))..)))))).)))))))))((((....((((..(((....)))..)))).....(((((....)))))))))..........................))))).)).)))....-- ( -45.90, z-score = -0.12, R) >droEre2.scaffold_4845 4343046 151 + 22589142 ---------AAG-AUUUCAAGU-UGCACUCCGUCGGCGCCAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUU-----UUAUUUCUCCUGUUGAAUUUUUCCUUUU-- ---------(((-(.(((((((-((((((((..(((.((((......)-))).)))..))...)))..)))))((((....((((..(((....)))..)))).....(((((....))))))))).........-----............))))).))))......-- ( -39.60, z-score = 0.11, R) >dp4.chr3 4732446 162 - 19779522 GCCACGUGGCCUAGUGUCAUGUAGGCACUUGGCCGACGUUAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUUG----CUCCUUCUUUUCUUUUUUUUUUCUCUU--- ((.((((((((.((((((.....)))))).)))).)))).....((((-(.(((.((((((((........))))((..(.((((..(((....)))..))))..)..))..)))).))).))))).........)----)..........................--- ( -53.70, z-score = -2.09, R) >droPer1.super_2 4925227 161 - 9036312 GCCACGUGGCCUAGUGUCAUGUAGGCACUUGGCCGACGUUAAGAACAU-GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUU------UUGCUCCUUCUGUUCUUUUUUUUCUCUU-- ...((((((((.((((((.....)))))).)))).)))).(((((((.-(((.((((..((((........))))......((((..(((....)))..)))).)))))))......((((((...........------.))))))...)))))))...........-- ( -55.90, z-score = -2.32, R) >droWil1.scaffold_180700 4631899 152 - 6630534 ---------------UUCAAAU-UGUGCUUUAGGCAACAGCGUAGCUCAAGCCUCUUUAGCUCAG-UGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAACUCAAAUUCUUA-AAUGAUAAUUUUUUCUUUUUUUUAGUUCUUUU ---------------.....((-((.(((..((((...(((...)))...))))....))).)))-)...(((((.(((((.......)))))(((...((((((....((((....))))..))))))...)))..-........................)))))... ( -34.80, z-score = -0.32, R) >droVir3.scaffold_12875 7716987 154 + 20611582 -GCAACGUGGCAGGUUUGAAGU-AAGCAUUUGGGUAUUGAUUGAGCAUAAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAUUUUUUUU-UUCUUCUAAAUUCUACUUUU------------- -((..((.((.(((((((((..-...(((..((((..((......))...))))..)))((((........))))......((((..(((....)))..)))))))))))))..))))..))..............-....................------------- ( -39.30, z-score = 0.50, R) >anoGam1.chr2R 57350968 142 - 62725911 ---------------UUAAAAU-AAUGCGUUGACGAGUGAGAAAGUAUGGGACUCCGUGGCGCAA-CGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAUCUUUUUUUUCUCAUUUUGACGAAAGCCACUCACUU----------- ---------------.......-....(((..(..((((((((((...(((((((((..((((..-.....)))).((((((....).))))).....(((((.......)))))))))))).))....)))))))))))..)))..............----------- ( -40.90, z-score = -0.63, R) >consensus GCCACGUGGACU_GUUUCAAGU_UGCGCUCCGACGACGCCAAGAACAU_GGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUU_____UUAUUACUUCUAUUGAAUUUUUCCUUUU__ ............................................((((..(((((((..((((........)))).((((.........)))).....(((((.......))))))))))))))))............................................ (-27.92 = -25.47 + -2.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:51 2011