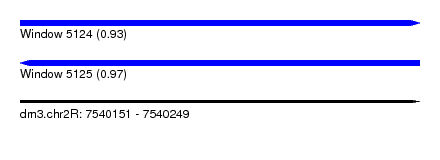
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,540,151 – 7,540,249 |
| Length | 98 |
| Max. P | 0.972572 |

| Location | 7,540,151 – 7,540,249 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Shannon entropy | 0.41648 |
| G+C content | 0.49109 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -27.22 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
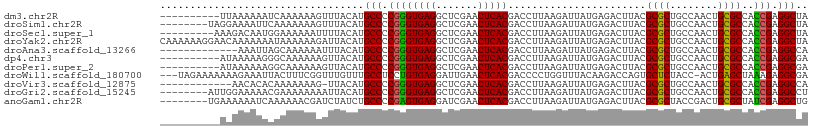
>dm3.chr2R 7540151 98 + 21146708 UAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAACUUUUUUGAUUUUUUAA---------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))........................---------- ( -29.10, z-score = -1.15, R) >droSim1.chr2R 6062887 100 + 19596830 UAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAACUUUUUUUGAAUUUUCCUA-------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))..........................-------- ( -29.10, z-score = -0.83, R) >droSec1.super_1 5090820 99 + 14215200 UAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAAAUUUUUUCCAUUGUCUUU--------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).))))))).........................--------- ( -29.10, z-score = -1.30, R) >droYak2.chr2R 11724664 108 - 21139217 UAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAUCUUUUUUAUUUUUUUGUUCCUUUUUUG ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).))))))).................................. ( -29.10, z-score = -1.36, R) >droAna3.scaffold_13266 8760069 95 + 19884421 UGGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAAUUUUUUGCUAAUUU------------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))..((((.....))))......------------- ( -29.20, z-score = -0.77, R) >dp4.chr3 4725703 98 + 19779522 UCGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUUGCCCUUUUUAU---------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))........................---------- ( -29.20, z-score = -0.62, R) >droPer1.super_2 4918447 98 + 9036312 UCGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAACUUUUUUGCCUUUUUUAU---------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))........................---------- ( -29.20, z-score = -0.77, R) >droWil1.scaffold_180700 4630805 104 + 6630534 UCGCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAACAAACCGAAAGUAAUUUCUUUUUUUCUA--- ..(((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).........((((....))))..........--- ( -30.60, z-score = -2.12, R) >droVir3.scaffold_12875 7707451 95 - 20611582 UGGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAA-CUUUUUUUGUGUGUU------------ ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))......-...............------------ ( -28.90, z-score = -0.38, R) >droGri2.scaffold_15245 7464591 100 + 18325388 AGGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAUUUUUUUUCGUUUUUCCAAU-------- ..(((((((((((((........))))((..(.((((..(((....)))..))))..)..))..)).)))))))..........................-------- ( -28.90, z-score = -1.06, R) >anoGam1.chr2R 57350466 100 + 62725911 CAGCCUCGAUAGCGCAGUCGGUAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAUCCUCACUCGGGGCAGAUAGAUCGUUUUUUGAUUUUUUCA-------- ..(((((((..((((........))))......((((..(((....)))..))))............))))))).((.((((((.....))))))..)).-------- ( -29.90, z-score = -2.07, R) >consensus UAGCCUCGGUGGCGCAGUUGGCAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAGCCUCACCCGGGGCAUGUAAACUUUUUUCCUUUUUUAU__________ ..(((((((((((((........))))......((((..(((....)))..)))).........)).))))))).................................. (-27.22 = -26.60 + -0.62)

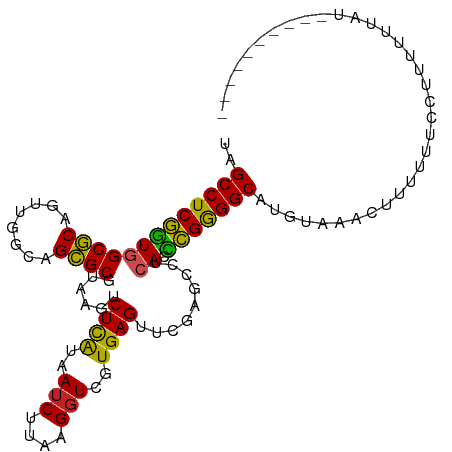
| Location | 7,540,151 – 7,540,249 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Shannon entropy | 0.41648 |
| G+C content | 0.49109 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.34 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7540151 98 - 21146708 ----------UUAAAAAAUCAAAAAAGUUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUA ----------........................(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -25.00, z-score = -1.66, R) >droSim1.chr2R 6062887 100 - 19596830 --------UAGGAAAAUUCAAAAAAAGUUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUA --------..........................(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -25.00, z-score = -0.90, R) >droSec1.super_1 5090820 99 - 14215200 ---------AAAGACAAUGGAAAAAAUUUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUA ---------.........................(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -25.00, z-score = -1.44, R) >droYak2.chr2R 11724664 108 + 21139217 CAAAAAAGGAACAAAAAAAUAAAAAAGAUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUA ..................................(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -25.00, z-score = -1.75, R) >droAna3.scaffold_13266 8760069 95 - 19884421 -------------AAAUUAGCAAAAAAUUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCCA -------------.....................(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -24.60, z-score = -1.45, R) >dp4.chr3 4725703 98 - 19779522 ----------AUAAAAAGGGCAAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCGA ----------.......................((((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).)))). ( -25.80, z-score = -0.48, R) >droPer1.super_2 4918447 98 - 9036312 ----------AUAAAAAAGGCAAAAAAGUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCGA ----------.......................((((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).)))). ( -25.80, z-score = -0.92, R) >droWil1.scaffold_180700 4630805 104 - 6630534 ---UAGAAAAAAAGAAAUUACUUUCGGUUUGUUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGCGA ---..((((..((....))..)))).......(((((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))))) ( -30.70, z-score = -2.64, R) >droVir3.scaffold_12875 7707451 95 + 20611582 ------------AACACACAAAAAAAG-UUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCCA ------------...............-......(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -24.60, z-score = -1.55, R) >droGri2.scaffold_15245 7464591 100 - 18325388 --------AUUGGAAAAACGAAAAAAAAUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCCU --------.(((......))).............(((.((((((((.((((.(.((..........)).).)))).)))))((((........))))..))).))).. ( -24.90, z-score = -1.51, R) >anoGam1.chr2R 57350466 100 - 62725911 --------UGAAAAAAUCAAAAAACGAUCUAUCUGCCCCGAGUGAGGAUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUACCGACUGCGCUAUCGAGGCUG --------..........................(((.((((((((.......)))))...((((......))))......((((........))))..))).))).. ( -23.00, z-score = -2.02, R) >consensus __________AAAAAAAAAGAAAAAAAUUUACAUGCCCCGGGUGAGGCUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUGCCAACUGCGCCACCGAGGCUA ..................................(((.((((((((.......))))).......................((((........))))..))).))).. (-23.72 = -23.34 + -0.39)
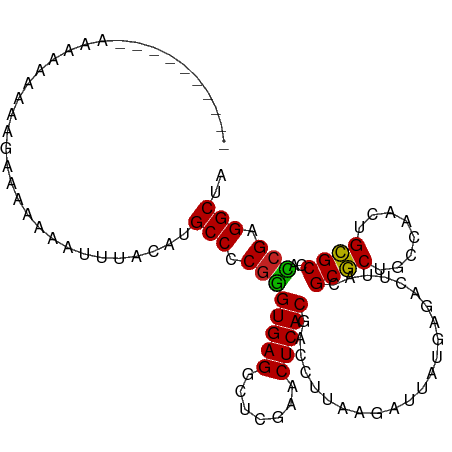
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:49 2011