| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,593,894 – 2,593,984 |
| Length | 90 |
| Max. P | 0.709979 |

| Location | 2,593,894 – 2,593,984 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.38930 |
| G+C content | 0.42988 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.16 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
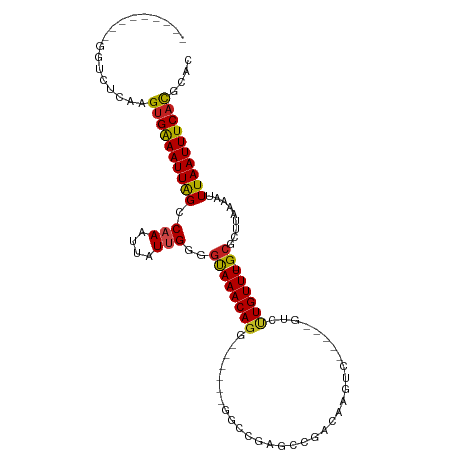
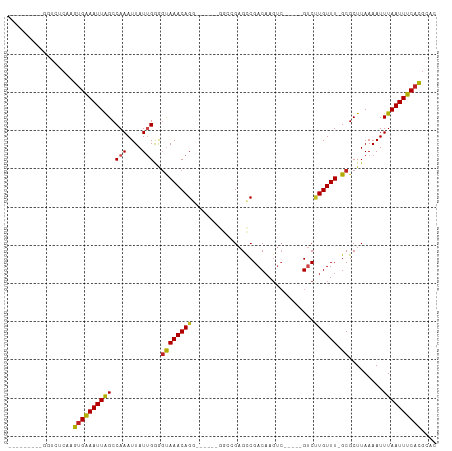
>dm3.chr2L 2593894 90 - 23011544 ---------GGUCUCAAGUGAAAUUAGCCAAAUUAUUGGGGUAAACAG-------GGCCGAGCCGACAAGUC-----GUCCUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ---------........((((((((((((((....)))).((((((((-------(((.((.(......)))-----)))))))))-)).........)))))))))).... ( -28.10, z-score = -2.95, R) >droSim1.chr2L 2552292 90 - 22036055 ---------GGUCUCAAGUGAAAUUAGCCAAAUUAUUGGGGUAAACAG-------GGCCGAGCCGACAAGUC-----GUCCUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ---------........((((((((((((((....)))).((((((((-------(((.((.(......)))-----)))))))))-)).........)))))))))).... ( -28.10, z-score = -2.95, R) >droSec1.super_5 762230 90 - 5866729 ---------GGUCUCAAGUGAAAUUAGCCAAAUUAUUGGGGUAAACAG-------GGCCGAGCCGACAAGUC-----GUCCUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ---------........((((((((((((((....)))).((((((((-------(((.((.(......)))-----)))))))))-)).........)))))))))).... ( -28.10, z-score = -2.95, R) >droYak2.chr2L 2581959 101 - 22324452 --------AAGUCUCAAGUGAAAUUGGCCAAAUUAUUGGGGUAAACAGG---UGAGGCAGAGCCGACAACUCGACUUGUCCUGUUUGGCGCUUAAAAUUUAAUUUCACGCAC --------.........((((((((((((((....))))(.((((((((---((((...(((.......)))..)))).)))))))).).........)))))))))).... ( -28.60, z-score = -1.72, R) >droEre2.scaffold_4929 2638553 91 - 26641161 ---------GGUCUCAAGUGAAAUUGGCCAAAUUAUUGGGUUAAACAGG------GGCCGAGCCGACAAGUC-----GUCCUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ---------........(((((((((....((((.((((((((((((((------(((...)))((....))-----..)))))))-).))))))))))))))))))).... ( -24.80, z-score = -1.32, R) >droAna3.scaffold_12913 405364 99 + 441482 -GCCCGAUGGCUCACAUGAGGAAUUAGCCAAAUUAUUGAAGUAAACAGG------AGGCGAGUCGACAAGUC-----GUCGUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC -(((....)))........((((((((.(((....)))..(((((((.(------((((..(....)..)))-----.)).)))))-)).........))))))))...... ( -20.50, z-score = 0.43, R) >dp4.chr4_group4 1898759 89 - 6586962 -----------UCCCAAGUGAAAUUAGCCAAAUUAUGGAGGCAAACAGG------GGCAGAGCCGACAAG-----UCGGCUUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC -----------......((((((((((((..........)))....(((------.((((((((((....-----)))))))...)-)).)))......))))))))).... ( -30.20, z-score = -3.58, R) >droPer1.super_10 906016 89 - 3432795 -----------UCCCAAGUGAAAUUAGCCAAAUUAUGGAGGCAAACAGG------GGCAGAGCCGACAAG-----UCGGCUUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC -----------......((((((((((((..........)))....(((------.((((((((((....-----)))))))...)-)).)))......))))))))).... ( -30.20, z-score = -3.58, R) >droWil1.scaffold_180772 1236190 103 - 8906247 ------UGUUAAGCAAAGUGAAAUUAGCCAAAUUAUUGGAGUAAACAGGAUGCACAGGAGAGUCGACAAGUC--AUCGUCUUGUUU-GCGCUUAAAAUUUAAUUUCAUGCAC ------......(((...(((((((((((((....)))).((((((((((((.((......)).((....))--..))))))))))-)).........)))))))))))).. ( -25.10, z-score = -1.92, R) >droVir3.scaffold_12963 11166008 98 - 20206255 GGGCCAAUGCUUCACUUGUGAAAUUAGCCAAAUUAUUGGAGUAAACAGG--------GCGAGUCGACAAGUC-----GUCUUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ((((....)))).....((((((((((((((....)))).(((((((((--------((((.(.....).))-----)))))))))-)).........)))))))))).... ( -30.80, z-score = -3.29, R) >droMoj3.scaffold_6500 19900382 98 - 32352404 GGGGCAAUGCUUCAGUUGUGAAAUUAGCCAAAUUAUUGGAGUAAACAGG--------GCGAGUCGACAAGUC-----GUCUUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC (((((...))))).((.((((((((((((((....)))).(((((((((--------((((.(.....).))-----)))))))))-)).........)))))))))))).. ( -33.80, z-score = -4.34, R) >droGri2.scaffold_15252 5300934 100 - 17193109 UUGGCAAUGCUUCACAUGUGAAAUUAGCCAAAUUAUUGGAGUAAACAGG------UCGAGAGUCGACUAGUC-----GUCUUGUUU-GCGCUUAAAAUUUAAUUUCACGCAC ...((.(((.....)))((((((((((((((....)))).(((((((((------.(((.((....))..))-----).)))))))-)).........)))))))))))).. ( -29.50, z-score = -3.02, R) >consensus _________GGUCUCAAGUGAAAUUAGCCAAAUUAUUGGGGUAAACAGG______GGCCGAGCCGACAAGUC_____GUCUUGUUU_GCGCUUAAAAUUUAAUUUCACGCAC .................((((((((((.(((....)))((((((((((................................))))))...)))).....)))))))))).... (-10.81 = -10.16 + -0.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:45 2011