
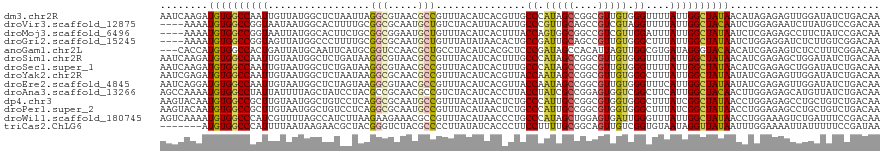
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,539,439 – 7,539,559 |
| Length | 120 |
| Max. P | 0.547032 |

| Location | 7,539,439 – 7,539,559 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.63242 |
| G+C content | 0.46544 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.01 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
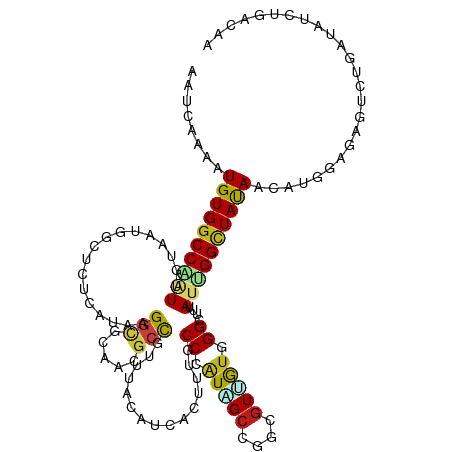
>dm3.chr2R 7539439 120 + 21146708 AAUCAAGAUGUGGCCAAUUGUUAUGGCUCUAAUUAGGCGUAACGCCGUUUACAUCACGUUGCCCAUAGCCGGCGUUGUGGGUUUUAUUGGCUAUAACAUAGAGAGUUGGAUAUCUGACAA ..(((.(((((..(((((((((((((((.......((((...))))..............(((((((((....)))))))))......))))))))))......)))))))))))))... ( -36.50, z-score = -1.60, R) >droVir3.scaffold_12875 7706725 116 - 20611582 ----AAAAUGUGGCCGGUAAUAAUGGCACUUUUGCGGCGCAAUGCUGUCUACAUUACAUUGCCCGUUGCAGCCGUCGUAGGUUUUAUUGGCUACAAUCUGGAGAAUCUUAUGUCCGACAA ----....((((((((((((.....(((.....((((.((((((.((....))...)))))))))))))((((......))))))))))))))))...((((.(......).)))).... ( -36.30, z-score = -1.52, R) >droMoj3.scaffold_6496 6699838 116 - 26866924 ----AAAAUGUGGCCGGUAAUUAUGGCACUUCUGCGGCGGAAUGCUGUUUACAUCACUUUACCAGUGGCGGCCGUCGUUGGAUUUAUUGGCUAUAAUCUCGAGAGCCUUCUAUCCGACAA ----.....(((((((((((..((((((.(((((...))))))))))))))).(((((.....))))))))))).)(((((((.....((((...........))))....))))))).. ( -37.50, z-score = -1.98, R) >droGri2.scaffold_15245 7463842 116 + 18325388 ----AAAAUGUGGCCGGUAGUUAUGGCCCUUUUGCGGCGCAAUGCUGUUUAUAUAACACUGCCGAUUGCAGCCGUUGUGGGCUUUAUUGGCUAUAAUCUGGAGGAUCUCUUGUCGGACAA ----....((((((((((((....(((((....((((((((((((((((.....))))..))..))))).)))))...))))))))))))))))).((((((((....))).)))))... ( -42.80, z-score = -2.74, R) >anoGam1.chr2L 30999840 117 + 48795086 ---CACCAUGUGGCCACUGAUUAUGCAAUUCAUGCGGUCCAACGCUGCCUACAUCACGCUCCCGAUAGCCACAUUAGUUGGCGUGAUAGGGUACAACAUCGAGAGUCUCCUUUCGGACAA ---..(((((..((..........))....)))).)((((...(.(((((..((((((((..(.............)..)))))))).))))))......(((((....))))))))).. ( -30.02, z-score = 0.15, R) >droSim1.chr2R 6062171 120 + 19596830 AAUCAAGAUGUGGCCAAUUGUAAUGGCUCUGAUAAGGCGUAACGCCGUUUACAUCACUUUGCCCAUAGCCGGCGUUGUGGGUUUUAUUGGCUAUAACAUCGAGAGCUGGAUAUCUGACAA ..(((.((((((((((((((((((((((((....)))......)))).))))).......(((((((((....)))))))))...)))))))....((.(....).)).))))))))... ( -33.70, z-score = -0.53, R) >droSec1.super_1 5090103 120 + 14215200 AAUCAAGAUGUGGCCAAUUGUAAUGGCUCUGAUAAGGCGUAACGCCGUUUACAUCACUUUGCCCAUAGCCGGCGUUGUGGGUUUUAUUGGCUAUAACAUCGAGAGCUGGAUAUCUGACAA ..(((.((((((((((((((((((((((((....)))......)))).))))).......(((((((((....)))))))))...)))))))....((.(....).)).))))))))... ( -33.70, z-score = -0.53, R) >droYak2.chr2R 11723950 120 - 21139217 AAUCGAGAUGUGGCCAAUUGUAAUGGCUCUAAUAAGGCGCAACGCCGUUUACAUCACGUUACCAAUAGCCGGCGUUGUGGGCUUUAUUGGCUAUAAUAUCGAGAGUUGGAUAUCUGACAA ..((((..((((((((((......((((((....)).((((((((((.(((..............))).))))))))))))))..))))))))))...))))..((..(....)..)).. ( -38.84, z-score = -2.28, R) >droEre2.scaffold_4845 4334669 120 - 22589142 AAUCAGGAUGUGGCCAAUUGUAAUGGCUCUAGUAAGGCGUAACGCCGUUUACAUCACGUUACCAAUAGCCGGCGUUGUGGGUUUCAUUGGCUAUAAUAUCGAGAGUUGGAUAUCUGACAA ..((((((((((((((.......))))....(((.((((...))))...)))..)))))).(((((...((..(((((((.(......).)))))))..))...)))))....))))... ( -34.20, z-score = -0.88, R) >droAna3.scaffold_13266 8759356 120 + 19884421 AGCCAAAAUGUGGCCUAUUAUUUUAGCUAUCCUACGCCGCAACGCCGUCUACAUCACCUUACCUAUCGCCGGAGUGGUCGGCUUCAUUGGCUACAACUUGGAGAGCAUGUUAUCUGACAA ..((((..(((((((..........((........))......((((.((((.((................)))))).))))......)))))))..))))......((((....)))). ( -28.69, z-score = 0.09, R) >dp4.chr3 4724997 120 + 19779522 AAGUACAAUGUGGCCGCUUGUAAUGGCUGUCCUCAGGCGCAAUGCCGUUUACAUAACUCUGCCCAUUGCCGGCGUGGUGGGCUUUAUCGGCUAUAACCUGGAGAGCCUGCUGUCUGACAA ..((....((((((((..(((((((((((((....))))....)))).))))).......((((((..(....)..)))))).....))))))))....(((.((....)).))).)).. ( -38.50, z-score = 0.18, R) >droPer1.super_2 4917741 120 + 9036312 AAGUACAAUGUGGCCGCUUGUAAUGGCUGUCCUCAGGCGCAAUGCCGUUUACAUAACUCUGCCCAUUGCCGGCGUGGUGGGCUUUAUCGGCUAUAACCUGGAGAGCCUGCUGUCUGACAA ..((....((((((((..(((((((((((((....))))....)))).))))).......((((((..(....)..)))))).....))))))))....(((.((....)).))).)).. ( -38.50, z-score = 0.18, R) >droWil1.scaffold_180745 2107195 120 - 2843958 AGUCAAAAUGUGGCCCAUCGUUUUAGCCAUCUUAAGAAGAAACGCCGUUUACAUAACCCUGCCCAUAGCUGGAGUGAUUGGGUUUAUUGGCUAUAACCUGGAAAGUCUGAUUUCCGACAA .(((....(((((((...((((((..(........)..))))))..........(((((.(((((....))).))....)))))....)))))))....(((((......)))))))).. ( -29.00, z-score = -0.57, R) >triCas2.ChLG6 4720935 113 + 13544221 -------AUGUGGCCCAUUUUAAUAAGAACGCUACGGGUCUACGCCCCUUAUAUCACCCUUCCUUUUGCGGCAGUUGUCGGUGUAAUAGGUUAUAAUUUGGAAAAUUAUUUUUCCGAUAA -------..(((((...((((....)))).)))))((((....))))....((((......(((.(((((.(.......).))))).))).........((((((....)))))))))). ( -24.80, z-score = -0.13, R) >consensus AAUCAAAAUGUGGCCAAUUGUAAUGGCUCUCAUAAGGCGCAACGCCGUUUACAUCACUUUGCCCAUAGCCGGCGUUGUGGGCUUUAUUGGCUAUAACAUGGAGAGUCUGAUAUCUGACAA ........((((((((((.................(((.....)))...............((.(((((....))))).))....))))))))))......................... (-15.90 = -16.01 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:47 2011