
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,511,678 – 7,511,784 |
| Length | 106 |
| Max. P | 0.978810 |

| Location | 7,511,678 – 7,511,784 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.44911 |
| G+C content | 0.56947 |
| Mean single sequence MFE | -41.29 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
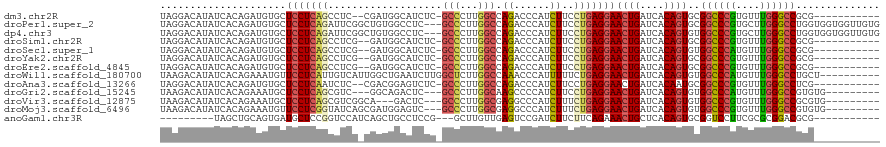
>dm3.chr2R 7511678 106 + 21146708 UAGGACAUAUCACAGAUGUGCUCCUCAGCCUC--CGAUGGCAUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCG----------- .....((((((...)))))).(((((((....--.(((((..(((.-(((...))).))).)))))...)))))))((((....))))((((((((....)))))))).----------- ( -45.10, z-score = -3.38, R) >droPer1.super_2 4890991 117 + 9036312 UAGGACAUAUCACAGAUGUGCUCCUCAGAUUCGGCUGUGGCCUC---GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGCUUGGGCCUGGUGGUGGUUGUG ((.(((.((((((......(((((((((....((.(.(((((..---......))))).).))......)))))))((((....))))))((((((....))))))..))))))))).)) ( -46.20, z-score = -0.94, R) >dp4.chr3 4698683 117 + 19779522 UAGGACAUAUCACAGAUGUGCUCCUCAGAUUCGGCUGUGGCCUC---GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCGUGCUUGGGCCUGGUGGUGGUUGUG ((.(((.((((((.(((..(.(((((((....((.(.(((((..---......))))).).))......))))))).)..))).......((((((....))))))..))))))))).)) ( -44.20, z-score = -0.50, R) >droSim1.chr2R 6034040 106 + 19596830 UAGGACAUAUCACAGAUGUGCUCCUCAGCCUCG--GAUGGCAUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCG----------- .....((((((...)))))).(((((((....(--(((((..(((.-(((...))).))).))))))..)))))))((((....))))((((((((....)))))))).----------- ( -46.80, z-score = -3.47, R) >droSec1.super_1 5062137 106 + 14215200 UAGGACAUAUCACAGAUGUGCUCCUCAGCCUCG--GAUGGCAUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCAUGUUUGGGCCGCG----------- .....((((((...)))))).(((((((....(--(((((..(((.-(((...))).))).))))))..)))))))((((....))))((((((((....)))))))).----------- ( -45.60, z-score = -3.29, R) >droYak2.chr2R 11695854 106 - 21139217 UAGGACAUAUCACAGAUGUGCUCCUCAGCCUCG--GAUGGCAUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCG----------- .....((((((...)))))).(((((((....(--(((((..(((.-(((...))).))).))))))..)))))))((((....))))((((((((....)))))))).----------- ( -46.80, z-score = -3.47, R) >droEre2.scaffold_4845 4305042 106 - 22589142 UAGGACAUAUCACAGAUGUGCUCCUCAGCCUCG--GAUGGCAUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCG----------- .....((((((...)))))).(((((((....(--(((((..(((.-(((...))).))).))))))..)))))))((((....))))((((((((....)))))))).----------- ( -46.80, z-score = -3.47, R) >droWil1.scaffold_180700 423912 110 + 6630534 UAAGACAUAUCACAGAAAUGUUCCUCAUUGUCAUUGGCUGAAUCUUGGCUCUUGGCCAAACCCAUUUUUCUGAGGAACUGAUCACAGUGUGGCCCAUGUUUGGGCCUGCU---------- ..............((...((((((((.......(((.......(((((.....)))))..)))......))))))))...))..((((.((((((....))))))))))---------- ( -32.72, z-score = -1.21, R) >droAna3.scaffold_13266 8735587 106 + 19884421 UAGGACAUAUCACAGAUGUGCUCCUCAAUCUC--CGACGGAGUCUC-GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAAUGCGGCCCGUGUUUGGGCCUCG----------- (((..((((((...)))))).((((((.....--.((.((.((((.-(((...))).)))))).))....)))))).)))........(.((((((....)))))).).----------- ( -34.90, z-score = -1.30, R) >droGri2.scaffold_15245 7433051 105 + 18325388 UAAGACAUAUCACAGAAAUGCUCCUCAGCGUC---GGCAGACUC---GCCCUUGGCAAGCCCCAUCUUCCUGAGGAACUGAUCACAGUGUGGCCCAUGUUUGGGCCGUGUG--------- ..........(((........(((((((.(((---....)))..---(((...))).............)))))))((((....))))..((((((....)))))))))..--------- ( -33.60, z-score = -0.90, R) >droVir3.scaffold_12875 7677447 105 - 20611582 UAAGACAUAUCACAGAAAUGCUCCUCAGCGUCGGCA---GACUC---GCCCUUGGCGAGGCCCAUCUUUCUGAGGAACUGAUCACAGUGUGGCCCGUGUUUGGGCCGCGUG--------- ..........(((........(((((((.((.(((.---..(((---(((...))))))))).))....)))))))((((....))))((((((((....)))))))))))--------- ( -42.10, z-score = -2.54, R) >droMoj3.scaffold_6496 6668500 108 - 26866924 UAAGACAUAUCACAGAAAUGUUCCUCGGUAUCAGCGAUGGAGUC---GCCCUUGGCGAGGCCCAUCUUUCUGAGGAACUGAUCACAGUGUGGCCCGUGUUUGGGCCGUGUG--------- ....((((..(((.((...(((((((((.......(((((..((---(((...)))))...)))))...)))))))))...))...))).((((((....)))))))))).--------- ( -41.50, z-score = -2.43, R) >anoGam1.chr3R 45909362 97 + 53272125 ---------UAGCUGCAGUGAUGCUCCGGUCCAUCAGCUGCCUCCG---GCUUGUUGAGUCCGAUCUUCUUCAGAAACUGCUCACAGUGCGGUCCUUCGCGCGGACGCG----------- ---------..((.(((....)))....((((...(((((....))---)))(((.((((....(((.....)))....)))))))((((((....)))))))))))).----------- ( -30.50, z-score = 0.32, R) >consensus UAGGACAUAUCACAGAUGUGCUCCUCAGCCUCG_CGAUGGCAUCUC_GCCCUUGGCCAGACCCAUCUUCCUGAGGAACUGAUCACAGUGCGGCCCGUGUUUGGGCCGCG___________ .....................(((((((...................(((...))).((......))..)))))))((((....))))..((((((....)))))).............. (-21.80 = -21.95 + 0.14)

| Location | 7,511,678 – 7,511,784 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.44911 |
| G+C content | 0.56947 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
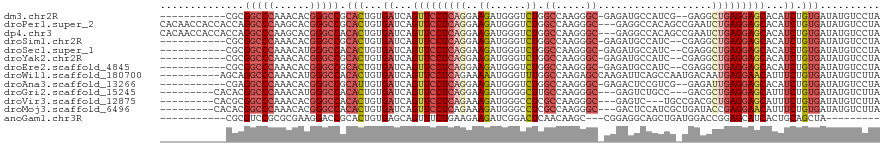
>dm3.chr2R 7511678 106 - 21146708 -----------CGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGAUGCCAUCG--GAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA -----------.(((((((......)))))))((...(((..(((((((((...(((((((((.(((...)))-.)))).))))).--....)))))))))..))).))........... ( -49.30, z-score = -3.11, R) >droPer1.super_2 4890991 117 - 9036312 CACAACCACCACCAGGCCCAAGCACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC---GAGGCCACAGCCGAAUCUGAGGAGCACAUCUGUGAUAUGUCCUA ((((..(((.....(((((......))))).....)))....((((((((((..(.(((((((.(((...)))---.))))).))..)...)))))))))).....)))).......... ( -47.60, z-score = -2.09, R) >dp4.chr3 4698683 117 - 19779522 CACAACCACCACCAGGCCCAAGCACGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC---GAGGCCACAGCCGAAUCUGAGGAGCACAUCUGUGAUAUGUCCUA ((((..(((.....(((((......))))).....)))....((((((((((..(.(((((((.(((...)))---.))))).))..)...)))))))))).....)))).......... ( -46.50, z-score = -2.03, R) >droSim1.chr2R 6034040 106 - 19596830 -----------CGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGAUGCCAUC--CGAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA -----------.(((((((......)))))))((...(((..(((((((((...(((((((((.(((...)))-.)))).)))))--.....)))))))))..))).))........... ( -48.20, z-score = -2.85, R) >droSec1.super_1 5062137 106 - 14215200 -----------CGCGGCCCAAACAUGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGAUGCCAUC--CGAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA -----------...((((((....)))))).(((...(((..(((((((((...(((((((((.(((...)))-.)))).)))))--.....)))))))))..))).))).......... ( -46.30, z-score = -2.51, R) >droYak2.chr2R 11695854 106 + 21139217 -----------CGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGAUGCCAUC--CGAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA -----------.(((((((......)))))))((...(((..(((((((((...(((((((((.(((...)))-.)))).)))))--.....)))))))))..))).))........... ( -48.20, z-score = -2.85, R) >droEre2.scaffold_4845 4305042 106 + 22589142 -----------CGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGAUGCCAUC--CGAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA -----------.(((((((......)))))))((...(((..(((((((((...(((((((((.(((...)))-.)))).)))))--.....)))))))))..))).))........... ( -48.20, z-score = -2.85, R) >droWil1.scaffold_180700 423912 110 - 6630534 ----------AGCAGGCCCAAACAUGGGCCACACUGUGAUCAGUUCCUCAGAAAAAUGGGUUUGGCCAAGAGCCAAGAUUCAGCCAAUGACAAUGAGGAACAUUUCUGUGAUAUGUCUUA ----------.(((((((((....)))))).(((...((...((((((((........((((((((.....)))))......)))........))))))))...)).)))...))).... ( -36.89, z-score = -2.37, R) >droAna3.scaffold_13266 8735587 106 - 19884421 -----------CGAGGCCCAAACACGGGCCGCAUUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC-GAGACUCCGUCG--GAGAUUGAGGAGCACAUCUGUGAUAUGUCCUA -----------...(((((......))))).(((...(((..(((((((((...(((((((((.(((...)))-.)))).))))).--....)))))))))..))).))).......... ( -45.90, z-score = -2.94, R) >droGri2.scaffold_15245 7433051 105 - 18325388 ---------CACACGGCCCAAACAUGGGCCACACUGUGAUCAGUUCCUCAGGAAGAUGGGGCUUGCCAAGGGC---GAGUCUGCC---GACGCUGAGGAGCAUUUCUGUGAUAUGUCUUA ---------.....((((((....)))))).(((...((...(((((((((...(.(((((((((((...)))---)))))..))---).).)))))))))...)).))).......... ( -43.60, z-score = -2.67, R) >droVir3.scaffold_12875 7677447 105 + 20611582 ---------CACGCGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGAAAGAUGGGCCUCGCCAAGGGC---GAGUC---UGCCGACGCUGAGGAGCAUUUCUGUGAUAUGUCUUA ---------.....(((((......))))).(((...((...(((((((((...(.(.(((((((((...)))---)))..---.))).)).)))))))))...)).))).......... ( -44.20, z-score = -2.96, R) >droMoj3.scaffold_6496 6668500 108 + 26866924 ---------CACACGGCCCAAACACGGGCCACACUGUGAUCAGUUCCUCAGAAAGAUGGGCCUCGCCAAGGGC---GACUCCAUCGCUGAUACCGAGGAACAUUUCUGUGAUAUGUCUUA ---------.....(((((......))))).(((...((...(((((((((...((((((..(((((...)))---)).)))))).))(....))))))))...)).))).......... ( -39.00, z-score = -2.80, R) >anoGam1.chr3R 45909362 97 - 53272125 -----------CGCGUCCGCGCGAAGGACCGCACUGUGAGCAGUUUCUGAAGAAGAUCGGACUCAACAAGC---CGGAGGCAGCUGAUGGACCGGAGCAUCACUGCAGCUA--------- -----------.((.((((((((......)))..((((((.....(((((......)))))))).))).).---)))).((((.(((((........))))))))).))..--------- ( -29.00, z-score = 0.98, R) >consensus ___________CGCGGCCCAAACACGGGCCGCACUGUGAUCAGUUCCUCAGGAAGAUGGGUCUGGCCAAGGGC_GAGAUGCCAUCG_CGAGGCUGAGGAGCACAUCUGUGAUAUGUCCUA ..............(((((......))))).(((...((...(((((((((..((......)).((.....))...................)))))))))...)).))).......... (-24.00 = -24.77 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:45 2011