
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,439,327 – 7,439,435 |
| Length | 108 |
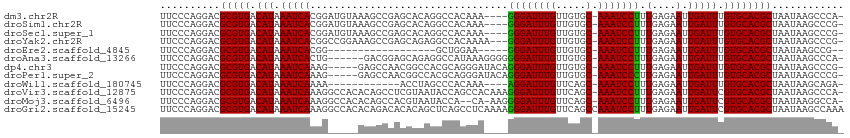
| Max. P | 0.996875 |

| Location | 7,439,327 – 7,439,435 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Shannon entropy | 0.48832 |
| G+C content | 0.46805 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -17.09 |
| Energy contribution | -16.94 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7439327 108 + 21146708 UUCCCAGGACGCGUGACAUAAAUCACGGAUGUAAAGCCGAGCACAGGCCACAAA----GGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCA- ..........(((((.(((((((((....(((...(((.......))).)))((----((((((((.....)-)))))))))......))))))))))))))...........- ( -34.60, z-score = -2.77, R) >droSim1.chr2R 5962991 108 + 19596830 UUCCCAGGACGCGUGACAUAAAUCACGGAUGUAAAGCCGAGCACAGGCCACAAA----GGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCG- ..........(((((.(((((((((....(((...(((.......))).)))((----((((((((.....)-)))))))))......))))))))))))))...........- ( -34.60, z-score = -2.85, R) >droSec1.super_1 4990586 108 + 14215200 UUCCCAGGACGCGUGACAUAAAUCACGGAUGUAAAGCCGAGCACAGGCCACAAA----GGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCG- ..........(((((.(((((((((....(((...(((.......))).)))((----((((((((.....)-)))))))))......))))))))))))))...........- ( -34.60, z-score = -2.85, R) >droYak2.chr2R 11618170 109 - 21139217 UUCCCAGGACGCGUGACAUAAAUCACGGCCGGAAAGCCGAGCAGAGGCCACAAAA---GGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCG- ..........(((((.(((((((((.((((.....((...))...)))).(..((---((((((((.....)-)))))))))..)...))))))))))))))...........- ( -37.70, z-score = -3.18, R) >droEre2.scaffold_4845 4230307 88 - 22589142 UUCCCAGGACGCGUGACAUAAAUCACGG------------------GCUGGAA-----GGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCG-- ..........(((((.(((((((((...------------------.((.(((-----((.(((((.....)-))))))))).))...))))))))))))))..........-- ( -31.10, z-score = -3.33, R) >droAna3.scaffold_13266 1153488 106 + 19884421 UUCCCAGGACGCGUGACAUAAAUCACUG------GACGGAGCAGAGGCCAUAAAGGGGGGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCA- ..........(((((.(((((((((.((------(.(........).)))......((((((((((.....)-)))))))))......))))))))))))))...........- ( -31.50, z-score = -1.02, R) >dp4.chr3 4625532 107 + 19779522 UUCCCAGGACGCGUGACAUAAAUCAAAG-----GAGCCAACGGCCACGCAGGGAUACAGGGAUUUGUUGUGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCG- ..........(((((.((((((((((.(-----..(((...)))..)..........(((((((((.....)-))))))))......)))))))))))))))...........- ( -33.50, z-score = -2.16, R) >droPer1.super_2 4812311 107 + 9036312 UUCCCAGGACGCGUGACAUAAAUCAAAG-----GAGCCAACGGCCACGCAGGGAUACAGGGAUUUGUUGUGC-AAAUCCCUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCG- ..........(((((.((((((((((.(-----..(((...)))..)..........(((((((((.....)-))))))))......)))))))))))))))...........- ( -36.20, z-score = -2.90, R) >droWil1.scaffold_180745 881269 96 - 2843958 UUCCCAGGACGCGUGACAUAAAUCAAAA------------ACCUAGCCCACAAA----AGGAUUUGUUCAGC-AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCAGA- ..........(((((.((((((((((..------------.....(....).((----((((((((.....)-))))))))).....)))))))))))))))...........- ( -29.10, z-score = -4.49, R) >droVir3.scaffold_12875 7587862 112 - 20611582 UUCCCAGGACGCGUGACAUAAAUCAAAGGCCACACAGCCUCGUAAUACCAGCCACAAAGGGAUUUGUUCAGC-AAAUCCUUUGAGAAUUGAUUCGUGCACGCUAAUAAGCCCA- ..........(((((.((..((((((((((......))))................((((((((((.....)-))))))))).....))))))..)))))))...........- ( -32.50, z-score = -3.38, R) >droMoj3.scaffold_6496 6576423 109 - 26866924 UUCCCAGGACGCGUGACAUAAAUCAAAGGCCACACAGCCACGUAAUACCA--CA-AAGGGGAUUUGUUCAGC-AAAUCCUUUGAGAAUUGAUUCGUGCACGCUAAUAAGGCCA- ..........(((((.((..((((((.(((......)))...........--(.-.((((((((((.....)-)))))))))..)..))))))..)))))))...........- ( -32.10, z-score = -2.89, R) >droGri2.scaffold_15245 7347380 114 + 18325388 UUCCCAGGACGCGUGACAUAAAUCAAAGGCCACACAGACACACAGCUCAGCCUCAAAAGGGAUUUGUUCAGCCAAAUCCUUUGAGAAUUGAUUCGUGCACGCUAAUAAGCCAAA ..........(((((.((..((((((.(((...................)))((..((((((((((......))))))))))..)).))))))..)))))))............ ( -30.41, z-score = -2.63, R) >consensus UUCCCAGGACGCGUGACAUAAAUCAAAG_____AAGCCGACCACAGGCCACAAA____GGGAUUUGUUGUGC_AAAUCCUUUGAGAAUUGAUUUGUGCACGCUAAUAAGCCCA_ ..........(((((.(((.(((((.................................((((((((.....).))))))).(....).))))).))))))))............ (-17.09 = -16.94 + -0.15)
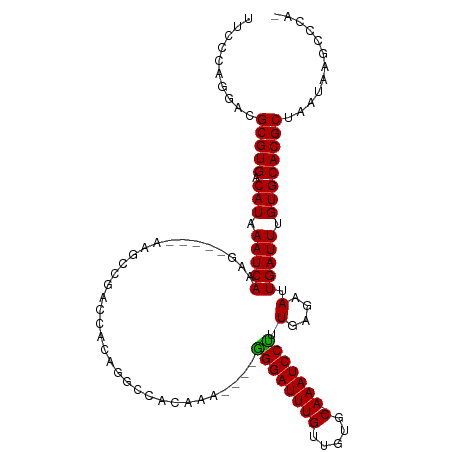

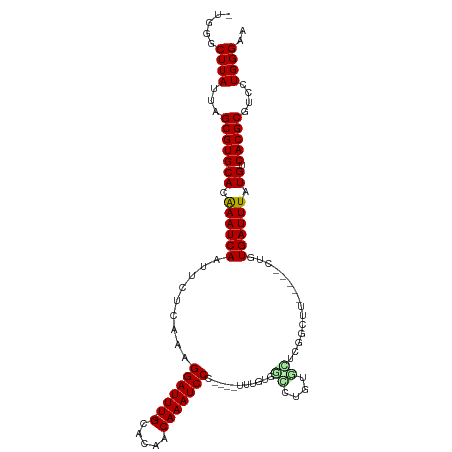
| Location | 7,439,327 – 7,439,435 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Shannon entropy | 0.48832 |
| G+C content | 0.46805 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7439327 108 - 21146708 -UGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCC----UUUGUGGCCUGUGCUCGGCUUUACAUCCGUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.((((((...(.((((((((((-(.....))))).))----)))).)(((.......)))..........)))))).)).)))))))))...... ( -31.00, z-score = -1.44, R) >droSim1.chr2R 5962991 108 - 19596830 -CGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCC----UUUGUGGCCUGUGCUCGGCUUUACAUCCGUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.((((((...(.((((((((((-(.....))))).))----)))).)(((.......)))..........)))))).)).)))))))))...... ( -31.30, z-score = -1.57, R) >droSec1.super_1 4990586 108 - 14215200 -CGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCC----UUUGUGGCCUGUGCUCGGCUUUACAUCCGUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.((((((...(.((((((((((-(.....))))).))----)))).)(((.......)))..........)))))).)).)))))))))...... ( -31.30, z-score = -1.57, R) >droYak2.chr2R 11618170 109 + 21139217 -CGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCC---UUUUGUGGCCUCUGCUCGGCUUUCCGGCCGUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.((((((..((.((((((((((-(.....))))))).---.)))).))........(((((....))))))))))).)).)))))))))...... ( -33.00, z-score = -1.50, R) >droEre2.scaffold_4845 4230307 88 + 22589142 --CGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCC-----UUCCAGC------------------CCGUGAUUUAUGUCACGCGUCCUGGGAA --..(((....)))(((((......(((((.....))))))-))))..........-----((((((.------------------(((((((....)))))).)..)))))). ( -23.70, z-score = -2.08, R) >droAna3.scaffold_13266 1153488 106 - 19884421 -UGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCCCCCCUUUAUGGCCUCUGCUCCGUC------CAGUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.((((((.........((((((-(.....)))))))....((..((((........)))).------.)))))))).)).)))))))))...... ( -26.20, z-score = -0.66, R) >dp4.chr3 4625532 107 - 19779522 -CGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCACAACAAAUCCCUGUAUCCCUGCGUGGCCGUUGGCUC-----CUUUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.(((((((........((((((-(.....)))))))............(..(((...)))..-----).))))))).)).)))))))))...... ( -29.80, z-score = -0.92, R) >droPer1.super_2 4812311 107 - 9036312 -CGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAGGGAUUU-GCACAACAAAUCCCUGUAUCCCUGCGUGGCCGUUGGCUC-----CUUUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((.(((((((......((((((((-(.....)))))))))..........(..(((...)))..-----).))))))).)).)))))))))...... ( -34.70, z-score = -2.04, R) >droWil1.scaffold_180745 881269 96 + 2843958 -UCUGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU-GCUGAACAAAUCCU----UUUGUGGGCUAGGU------------UUUUGAUUUAUGUCACGCGUCCUGGGAA -....((((...(((((((.(((((((..((((((((((((-(.....))))))))----)...))))......------------..))))))).)).)))))....)))).. ( -26.20, z-score = -1.89, R) >droVir3.scaffold_12875 7587862 112 + 20611582 -UGGGCUUAUUAGCGUGCACGAAUCAAUUCUCAAAGGAUUU-GCUGAACAAAUCCCUUUGUGGCUGGUAUUACGAGGCUGUGUGGCCUUUGAUUUAUGUCACGCGUCCUGGGAA -.((((......(((((((..(((((..((.((((((((((-(.....))))).)))))).))..........((((((....)))))))))))..)).)))))))))...... ( -36.20, z-score = -1.97, R) >droMoj3.scaffold_6496 6576423 109 + 26866924 -UGGCCUUAUUAGCGUGCACGAAUCAAUUCUCAAAGGAUUU-GCUGAACAAAUCCCCUU-UG--UGGUAUUACGUGGCUGUGUGGCCUUUGAUUUAUGUCACGCGUCCUGGGAA -...(((.....(((((((..((((((.((.((((((((((-(.....)))))))..))-))--.))........((((....)))).))))))..)).))))).....))).. ( -34.50, z-score = -2.18, R) >droGri2.scaffold_15245 7347380 114 - 18325388 UUUGGCUUAUUAGCGUGCACGAAUCAAUUCUCAAAGGAUUUGGCUGAACAAAUCCCUUUUGAGGCUGAGCUGUGUGUCUGUGUGGCCUUUGAUUUAUGUCACGCGUCCUGGGAA .....((((...(((((((..((((((.((((((((((((((......)))))))..)))))))....((((..(....)..))))..))))))..)).)))))....)))).. ( -35.70, z-score = -1.89, R) >consensus _UGGGCUUAUUAGCGUGCACAAAUCAAUUCUCAAAGGAUUU_GCACAACAAAUCCC____UUUGUGGCCUGUGCUCGGCUU_____CUGUGAUUUAUGUCACGCGUCCUGGGAA .....((((...(((((((.((((((.........((((((........))))))...........((....))...............)))))).)).)))))....)))).. (-17.37 = -17.42 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:35 2011