
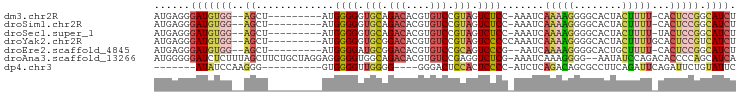
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,408,969 – 7,409,053 |
| Length | 84 |
| Max. P | 0.869403 |

| Location | 7,408,969 – 7,409,053 |
|---|---|
| Length | 84 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.56060 |
| G+C content | 0.53736 |
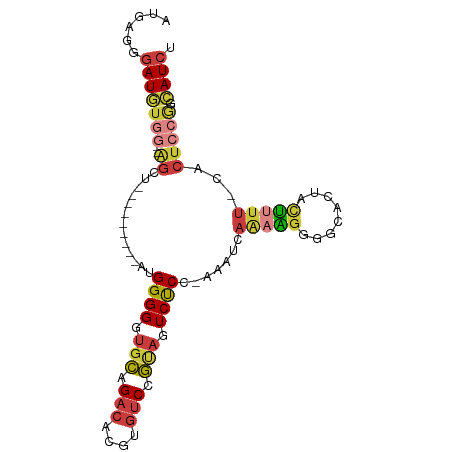
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7408969 84 + 21146708 AUGAGGGAUGUGG--AGCU---------AUGGGGGUGCAGACACGUGUCCGUAGUCUCC-AAAUCAAAAGGGGCACUACUUUU-CACUCCGGCAUCU .....((((((((--((..---------....(((..(......)..)))(((((((((-.........)))).)))))....-..)))).)))))) ( -27.80, z-score = -1.24, R) >droSim1.chr2R 5931605 84 + 19596830 AUGAGGGAUGUGG--AGCU---------AUGGGGGUGCAGACACGUGUCCGUAGUCUCC-AAAUCAAAAGGGGCACUACUUUU-CACUCCGGCAUCU .....((((((((--((..---------....(((..(......)..)))(((((((((-.........)))).)))))....-..)))).)))))) ( -27.80, z-score = -1.24, R) >droSec1.super_1 4959950 84 + 14215200 AUGAGGGAUGUGG--AGCU---------AUGGGGGUGCAGACACGUGUCCGUAGUCUCC-AAAUCAAAAGGGGCACUACUUUU-UACUCCGGCAUCU .....((((((((--((..---------....(((..(......)..)))(((((((((-.........)))).)))))....-..)))).)))))) ( -27.80, z-score = -1.42, R) >droYak2.chr2R 11586457 86 - 21139217 AUGAGGGAUGUGG--AGCU---------AUGGGGGUGCGGACACGUGUCCGUAGUCCCCCAAAUCAAAAGGGGCACUACUUUUGCACUCCGUCAUCU .....((((((((--((..---------.(((((((((((((....)))))))..))))))...((((((........))))))..))))).))))) ( -38.60, z-score = -3.47, R) >droEre2.scaffold_4845 4199108 83 - 22589142 AUGAGGGAUGUGG--AGCU---------AUGGGGAUGCGGACACGUGUCCGCAGUCCCG--AAUCAAAAGGGGCACUGCUUUU-CACUCCGGCAUCU .....((((((((--((..---------.((..(((((((((....)))))))(((((.--........))))).....))..-)))))).)))))) ( -35.20, z-score = -2.68, R) >droAna3.scaffold_13266 1120173 94 + 19884421 AUGGGGGAUCUCUUUAGCUUCUGCUAGGAGGGGGUGGCAGACACGUGUCCGAGGUCUCG-AAAUCAAAGGGG--AAUAUCCAGACACCCCAGCAUCA .(((((((((((..((((....))))(((.(.(.((.....))).).)))))))))...-........(((.--....))).....)))))...... ( -29.70, z-score = 0.06, R) >dp4.chr3 4592668 75 + 19779522 -------AUAUCCAAGGG----------GUGGGGUUGGGG----GGGACUCCACUCCCC-AUCUCAGACAGCGCCUUCAGAUUCAGAUUCUGUAUUC -------........(((----------((((((.(((((----....)))))..))))-)))))............((((.......))))..... ( -29.60, z-score = -1.74, R) >consensus AUGAGGGAUGUGG__AGCU_________AUGGGGGUGCAGACACGUGUCCGUAGUCUCC_AAAUCAAAAGGGGCACUACUUUU_CACUCCGGCAUCU ......(((((...................((((.(((.(((....))).))).))))...........((((.............)))).))))). (-14.03 = -14.52 + 0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:32 2011