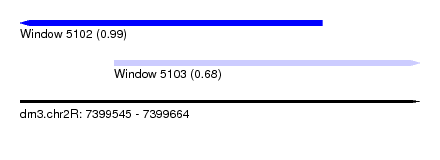
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,399,545 – 7,399,664 |
| Length | 119 |
| Max. P | 0.985596 |

| Location | 7,399,545 – 7,399,635 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.66 |
| Shannon entropy | 0.46944 |
| G+C content | 0.46492 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
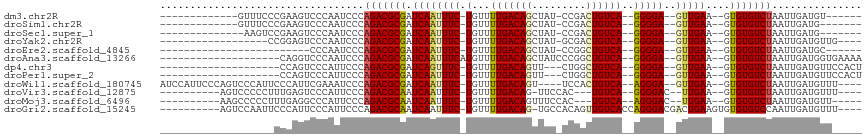
>dm3.chr2R 7399545 90 - 21146708 -------------GUUUCCCGAAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAU-CCGACUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGU------ -------------..............((((...(((((((.((((((((-(...(((((((....-....))))))--)))))--))))).--.))))))).))))....------ ( -25.40, z-score = -1.80, R) >droSim1.chr2R 5922289 89 - 19596830 -------------GUUUCCCGAAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAU-CCGACUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUG------- -------------..............((((...(((((((.((((((((-(...(((((((....-....))))))--)))))--))))).--.))))))).))))...------- ( -25.40, z-score = -1.99, R) >droSec1.super_1 4950678 88 - 14215200 --------------AAGUCCGAAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAU-CCGACUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUG------- --------------.............((((...(((((((.((((((((-(...(((((((....-....))))))--)))))--))))).--.))))))).))))...------- ( -25.40, z-score = -1.98, R) >droYak2.chr2R 11575997 87 + 21139217 ------------------CCGGAGUCCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAU-GCGACUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGUUG---- ------------------..(((.......))).(((((((.((((((((-(...(((((((....-....))))))--)))))--))))).--.)))))))...........---- ( -26.50, z-score = -1.73, R) >droEre2.scaffold_4845 4190093 78 + 22589142 -------------------------CCCAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGCUAU-CCGGCUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGC------ -------------------------..((((...(((((((.((((((((-(...(((((((((..-..))))))))--)))))--))))).--.))))))).))))....------ ( -30.10, z-score = -3.91, R) >droAna3.scaffold_13266 1109350 91 - 19884421 --------------------CAGGUCCCAAUCCCAGACGCGAUCAAUUUCAUGUUUUGACAGCUAUCCCGGCUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGGUGAAAA --------------------....(((((.((..(((((((.((((((((....((((((((((.....))))))))--)))))--))))).--.)))))))....))))).))... ( -31.00, z-score = -2.58, R) >dp4.chr3 4582783 87 - 19779522 --------------------CCAGUCCCAUUCCCAGACGCGAUCAGUUUC-UGUUUUGACAGUU---CUGGCUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGUUCCACU --------------------.(((((((.....((((.((.....)).))-))..((((((((.---...)))))))--)))))--.))).(--(((((((.....))))...)))) ( -24.60, z-score = -0.98, R) >droPer1.super_2 4770554 87 - 9036312 --------------------CCAGUCCCAUUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGUU---CUGGCUGUCA--GGGGA--GUUGAA--GUGUGUCUAAUUGAUGUUCCACU --------------------.......((((...(((((((.((((((((-(...((((((((.---...)))))))--)))))--))))).--.)))))))....))))....... ( -27.30, z-score = -2.05, R) >droWil1.scaffold_180745 826129 102 + 2843958 AUCCAUUCCCAGUCCCAUUCCCAUUCGAAAUCCCAGACGCGAUCAAUUUC-UGUUUUGACAGU----UCCACUGUCA--AGGGA--GUUGAA--GUGUGUCUAAUUGAUGUUU---- .........((((....(((......))).....(((((((.((((((((-(...(((((((.----....))))))--)))))--))))).--.))))))).))))......---- ( -25.80, z-score = -2.45, R) >droVir3.scaffold_12875 7540556 92 + 20611582 ----------AGUCCCCCUUUGAGUCCCAUUCCCAGACGCAAUCAAUUUC-UGUUUUGACAG-UUCCAC---UGUCA--GGGGAC--UUGAA--GUGUGUCUAAUUGAUGUUU---- ----------((.(.(.(((..((((((.....((((...........))-))..(((((((-.....)---)))))--))))))--)..))--).).).))...........---- ( -27.70, z-score = -2.84, R) >droMoj3.scaffold_6496 6523171 92 + 26866924 ----------AAGCCCCCUUUGAGGCCCAUUCCCAGACGCAAUCAAUUUC-UGUUUUGACAGUUUCCAC---UGUCA--AGGGAC--UUGAA--GUGUGUCUAAUUGAUGUU----- ----------..(((........))).((((...(((((((.((((.(((-(...((((((((....))---)))))--))))).--)))).--.)))))))....))))..----- ( -26.90, z-score = -2.60, R) >droGri2.scaffold_15245 7294187 101 - 18325388 ----------AGUCCAAUUCCCAUUCCCAUUCCCAGACGCAAUCAAUUUC-UGUUUUGACAG-UGCCACAGUUGUCACCAGGGACGACUGAAGUGUGUGUCCAAUUGAUGUUU---- ----------.................((((....((((((........(-(((....))))-.....((((((((......)))))))).....)))))).....))))...---- ( -21.60, z-score = -0.21, R) >consensus ___________________CCAAGUCCCAAUCCCAGACGCGAUCAAUUUC_UGUUUUGACAGCUAU_CCGACUGUCA__GGGGA__GUUGAA__GUGUGUCUAAUUGAUGUU_____ ..................................(((((((.(((((.........((((((.........)))))).........)))))....)))))))............... (-16.74 = -16.86 + 0.12)

| Location | 7,399,573 – 7,399,664 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 73.42 |
| Shannon entropy | 0.53497 |
| G+C content | 0.50740 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -13.41 |
| Energy contribution | -12.61 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
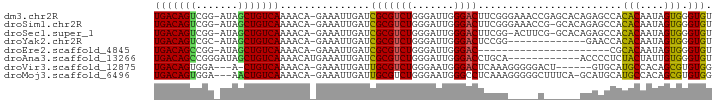
>dm3.chr2R 7399573 91 + 21146708 UGACAGUCGG-AUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUUCGGGAAACCGAGCACAGAGCCACACAAUAGUGGGUGU (((((((...-...))))))).....-..........((((((.......)))).((((....)))))).....(((.(((....)))))).. ( -28.20, z-score = -1.82, R) >droSim1.chr2R 5922316 90 + 19596830 UGACAGUCGG-AUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUUCGGGAAACCG-GCACAGAGCCACACAAUAGUGGGUGU (((((((...-...))))))).....-..........((((((.......))))..(((....)))-)).....(((.(((....)))))).. ( -26.50, z-score = -1.21, R) >droSec1.super_1 4950705 89 + 14215200 UGACAGUCGG-AUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUUCGG-ACUUCG-GCACAGAGCCACACAAUAGUGGGUGU .....(((((-(..((.(((((....-....))))).))(((((((........)))))-))))))-)).....(((.(((....)))))).. ( -24.90, z-score = -0.70, R) >droYak2.chr2R 11576027 78 - 21139217 UGACAGUCGC-AUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGACUCCGG-------------GAACCACACAAUAGUGGGUGU (((((((...-...))))))).....-.........(((.(((.(((.......))).)-------------)).((((......))))))). ( -21.80, z-score = -0.77, R) >droEre2.scaffold_4845 4190121 69 - 22589142 UGACAGCCGG-AUAGCUGUCAAAACA-GAAAUUGAUCGCGUCUGGGAUUGGGAC----------------------CGCACAAUAGUGGGUGU (((((((...-...))))))).....-....(..(((.(....).)))..).((----------------------(.(((....)))))).. ( -20.20, z-score = -1.27, R) >droAna3.scaffold_13266 1109384 81 + 19884421 UGACAGCCGGGAUAGCUGUCAAAACAUGAAAUUGAUCGCGUCUGGGAUUGGGACCUGCA------------ACCCCUCUACUAUUGUGGGUGU ...(((((.((((.((.(((((.........))))).)))))).)).)))..(((..((------------(...........)))..))).. ( -21.00, z-score = 0.20, R) >droVir3.scaffold_12875 7540586 82 - 20611582 UGACAGUGGA---A-CUGUCAAAACA-GAAAUUGAUUGCGUCUGGGAAUGGGACUCAAAGGGGGACU------GUGCAUGCCACAGCGUGUGG ((((((....---.-)))))).....-..........(((.(((((.((((..(((....)))..))------)).....)).)))))).... ( -22.10, z-score = -0.62, R) >droMoj3.scaffold_6496 6523200 88 - 26866924 UGACAGUGGA---AACUGUCAAAACA-GAAAUUGAUUGCGUCUGGGAAUGGGCCUCAAAGGGGGCUUUCA-GCAUGCAUGCCACAGCGUGUGG (((((((...---.))))))).....-.........((((((((((....((((((....))))))))))-).)))))..((((.....)))) ( -31.70, z-score = -2.39, R) >consensus UGACAGUCGG_AUAGCUGUCAAAACA_GAAAUUGAUCGCGUCUGGGAUUGGGACUUCGG_____C______AGAGCCACACAAUAGUGGGUGU (((((((.......)))))))...............(((((((.......))))........................(((....))).))). (-13.41 = -12.61 + -0.80)
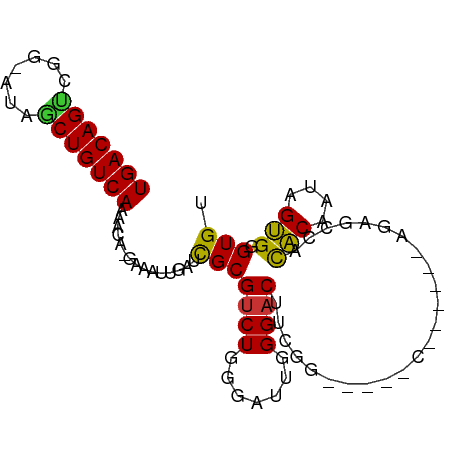
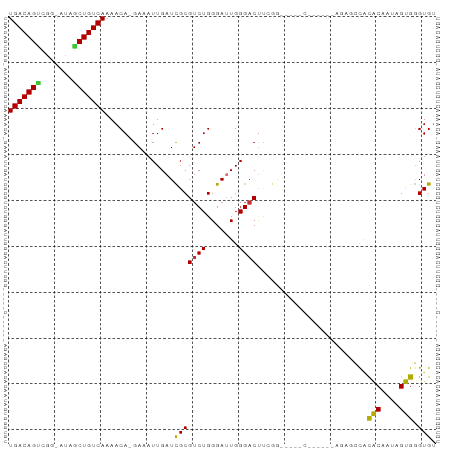
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:30 2011