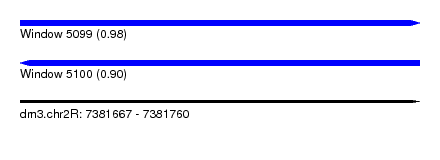
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,381,667 – 7,381,760 |
| Length | 93 |
| Max. P | 0.975809 |

| Location | 7,381,667 – 7,381,760 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.75 |
| Shannon entropy | 0.28433 |
| G+C content | 0.33085 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
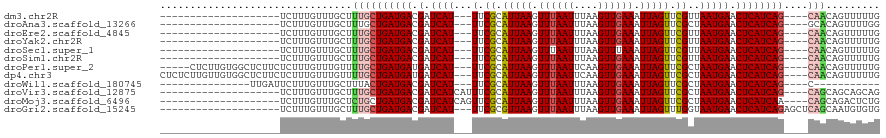
>dm3.chr2R 7381667 93 + 21146708 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGUUAAUGAACUCAUCAG----CAACAGUUUUUG --------------------.........(((((((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))).)))..... ( -26.10, z-score = -3.88, R) >droAna3.scaffold_13266 1092552 93 + 19884421 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG----GCACAGUUUUGG --------------------.........(((.(((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))).----))).)))..... ( -22.50, z-score = -2.03, R) >droEre2.scaffold_4845 4172656 93 - 22589142 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGUUAAUGAACUCAUCAG----CAACAGUUUUUG --------------------.........(((((((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))).)))..... ( -26.10, z-score = -3.88, R) >droYak2.chr2R 11557525 93 - 21139217 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGUUAAUGAACUCAUCAG----CAACAGUUUUUG --------------------.........(((((((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))).)))..... ( -26.10, z-score = -3.88, R) >droSec1.super_1 4932879 93 + 14215200 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUUAAAUUAGUUCGUUAAUGAACUCAUCAG----CAACAGUUUUUG --------------------.........(((((((((((((.(.((((---(.((.(((((.(((((.......)))))))))).))..))))).))))))))----))).)))..... ( -23.80, z-score = -3.63, R) >droSim1.chr2R 5904859 93 + 19596830 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGUUAAUGAACUCAUCAG----CAACAGUUUUUG --------------------.........(((((((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))).)))..... ( -26.10, z-score = -3.88, R) >droPer1.super_2 4752918 108 + 9036312 -----CUCUUGUGGCUCUUCUCUUUGUUUGUUUUGCUGAUGAUGAUCAU---UUCGCAUUAAGUUUAAUUCAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG----CAACAGUUUUUG -----......................((((..(((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))))))...... ( -25.00, z-score = -1.97, R) >dp4.chr3 4564049 113 + 19779522 CUCUCUUGUUGUGGCUCUUCUCUUUGUUUGUUUUGCUGAUGAUGAUCAU---UUCGCAUUAAGUUUAAUUCAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG----CAACAGUUUUUG ...........................((((..(((((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----))))))...... ( -25.00, z-score = -1.67, R) >droWil1.scaffold_180745 799439 87 - 2843958 ---------------UUGAUUCUUUGUUUGCUUUACUGAUGACGAUCAU---UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG----C----------- ---------------....................(((((((.(.((((---(.((.(((((.((((((....)))))).))))).))..))))).))))))))----.----------- ( -19.00, z-score = -1.91, R) >droVir3.scaffold_12875 7520219 96 - 20611582 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAUCAUUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG----CAGCAGCAGCAG --------------------....((((.(((((((((((((.(.((((.....((.(((((.((((((....)))))).))))).))...)))).))))))))----))).))))))). ( -28.40, z-score = -2.94, R) >droMoj3.scaffold_6496 6496648 96 - 26866924 --------------------UCUUUGUUUGCUCUGCUGAUGACGAUCAUCAGUUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAA----CAGCAGACUCUG --------------------.....(((((((..(((((((.....)))))))....................(((((....((((((....))))))..))))----)))))))).... ( -25.40, z-score = -2.43, R) >droGri2.scaffold_15245 7269588 97 + 18325388 --------------------UCUUUGUUUGCUUUGCUGAUGACGAUCAU---UUCGCGUUAAGUUUAAUUUAAGUUGAAAUUAGUUUGGUAAUGAACUCAUCAGAGCUCAGCAAUGUGUG --------------------.......(((((...(((((((.(.((((---((((..((((.((((((....)))))).))))..))).))))).)))))))).....)))))...... ( -22.80, z-score = -1.16, R) >consensus ____________________UCUUUGUUUGCUUUGCUGAUGACGAUCAU___UUCGCAUUAAGUUUAAUUUAAGUUGAAAUUAGUUCGCUAAUGAACUCAUCAG____CAACAGUUUUUG .......................(((((.......(((((((.(.((((.....((.(((((.((((((....)))))).))))).))...)))).)))))))).....)))))...... (-16.26 = -16.54 + 0.28)

| Location | 7,381,667 – 7,381,760 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Shannon entropy | 0.28433 |
| G+C content | 0.33085 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
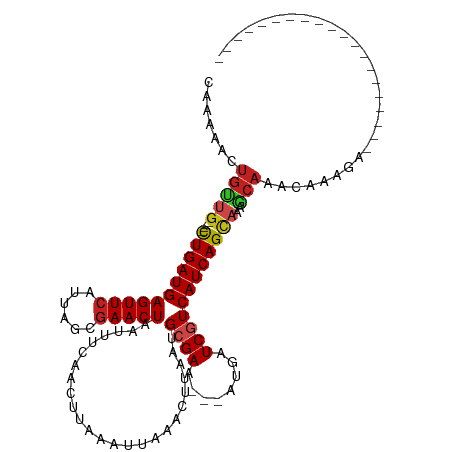
>dm3.chr2R 7381667 93 - 21146708 CAAAAACUGUUG----CUGAUGAGUUCAUUAACGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .........(((----(((((((((((......))))).........................((((.---....)))))))))))))............-------------------- ( -19.40, z-score = -2.77, R) >droAna3.scaffold_13266 1092552 93 - 19884421 CCAAAACUGUGC----CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- ......((.(((----.(((((((.(((((.(((((.....))).......((((....))))))..)---)))).).))))))))).))..........-------------------- ( -16.10, z-score = -1.34, R) >droEre2.scaffold_4845 4172656 93 + 22589142 CAAAAACUGUUG----CUGAUGAGUUCAUUAACGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .........(((----(((((((((((......))))).........................((((.---....)))))))))))))............-------------------- ( -19.40, z-score = -2.77, R) >droYak2.chr2R 11557525 93 + 21139217 CAAAAACUGUUG----CUGAUGAGUUCAUUAACGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .........(((----(((((((((((......))))).........................((((.---....)))))))))))))............-------------------- ( -19.40, z-score = -2.77, R) >droSec1.super_1 4932879 93 - 14215200 CAAAAACUGUUG----CUGAUGAGUUCAUUAACGAACUAAUUUAAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .........(((----(((((((((((......))))(((((((....))))))).............---.......))))))))))............-------------------- ( -19.50, z-score = -3.08, R) >droSim1.chr2R 5904859 93 - 19596830 CAAAAACUGUUG----CUGAUGAGUUCAUUAACGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .........(((----(((((((((((......))))).........................((((.---....)))))))))))))............-------------------- ( -19.40, z-score = -2.77, R) >droPer1.super_2 4752918 108 - 9036312 CAAAAACUGUUG----CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUGAAUUAAACUUAAUGCGAA---AUGAUCAUCAUCAGCAAAACAAACAAAGAGAAGAGCCACAAGAG----- .........(((----(((((((..(((((.(((.......(((.....)))..........)))..)---))))...))))))))))...........................----- ( -20.93, z-score = -1.83, R) >dp4.chr3 4564049 113 - 19779522 CAAAAACUGUUG----CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUGAAUUAAACUUAAUGCGAA---AUGAUCAUCAUCAGCAAAACAAACAAAGAGAAGAGCCACAACAAGAGAG .........(((----(((((((..(((((.(((.......(((.....)))..........)))..)---))))...))))))))))................................ ( -20.93, z-score = -1.60, R) >droWil1.scaffold_180745 799439 87 + 2843958 -----------G----CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA---AUGAUCGUCAUCAGUAAAGCAAACAAAGAAUCAA--------------- -----------(----((((((((.(((((.(((((.....))).......((((....))))))..)---)))).).))))))))...................--------------- ( -15.80, z-score = -1.72, R) >droVir3.scaffold_12875 7520219 96 + 20611582 CUGCUGCUGCUG----CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAAAUGAUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- .((((..(((((----.((((((..(((((.(((((.....))).......((((....))))))..)))))...)))))).))))).))))........-------------------- ( -25.90, z-score = -2.78, R) >droMoj3.scaffold_6496 6496648 96 + 26866924 CAGAGUCUGCUG----UUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAACUGAUGAUCGUCAUCAGCAGAGCAAACAAAGA-------------------- .....(((((((----.((((((..(((((((.(((.....))).......((((....)))).....))))))))))))).)))))))...........-------------------- ( -24.30, z-score = -1.93, R) >droGri2.scaffold_15245 7269588 97 - 18325388 CACACAUUGCUGAGCUCUGAUGAGUUCAUUACCAAACUAAUUUCAACUUAAAUUAAACUUAACGCGAA---AUGAUCGUCAUCAGCAAAGCAAACAAAGA-------------------- ......(((((..(((.((((((.(((..........((((((......))))))..........)))---....))))))..)))..))))).......-------------------- ( -16.45, z-score = -1.48, R) >consensus CAAAAACUGUUG____CUGAUGAGUUCAUUAGCGAACUAAUUUCAACUUAAAUUAAACUUAAUGCGAA___AUGAUCGUCAUCAGCAAAGCAAACAAAGA____________________ .......((((.....(((((((((((......))))).........................((((........))))))))))...))))............................ (-13.48 = -13.78 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:28 2011