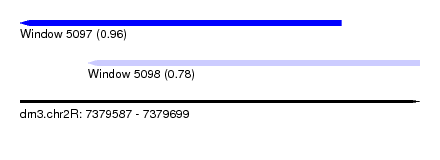
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,379,587 – 7,379,699 |
| Length | 112 |
| Max. P | 0.961879 |

| Location | 7,379,587 – 7,379,677 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.12 |
| Shannon entropy | 0.49300 |
| G+C content | 0.53460 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.89 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961879 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
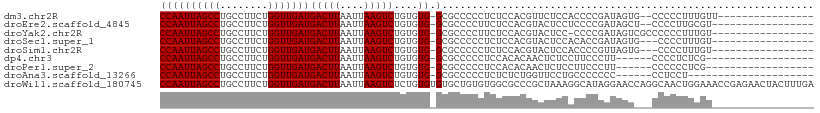
>dm3.chr2R 7379587 90 - 21146708 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCACGUUCUCCACCCCGAUAGUG--CCCCCUUUGUU---------------- ...(((((((........)))))))(((((....))))).....(-((((...........................)))--)).........---------------- ( -15.23, z-score = -0.68, R) >droEre2.scaffold_4845 4170651 89 + 22589142 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCUUCUCCACGUACUCCUCCCCGAUAGCU--CCCCUUGCGU----------------- ...(((((((........)))))))(((((....)))))...(((-(.(......).))))................((.--......))..----------------- ( -16.20, z-score = -0.77, R) >droYak2.chr2R 11555513 90 + 21139217 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCUUCUCCACGUACUCC-CCCCGAUAGUCGCCCCCCUUUGU----------------- ...(((((((........)))))))(((((....)))))...(((-(.(......).)))).......-.......................----------------- ( -14.80, z-score = -0.61, R) >droSec1.super_1 4930890 88 - 14215200 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCACGUACUCCACACCGAUAGUG---CCCCUUUGU----------------- ...(((((((........)))))))(((((....))))).(((((-(.(..(.........)..).))))))........---.........----------------- ( -17.60, z-score = -0.93, R) >droSim1.chr2R 5902932 88 - 19596830 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCACGUACUCCACCCCGUUAGUG---CCCCUUUGU----------------- ...(((((((........)))))))(((((....)))))...(((-(.(......).))))(((((..........))))---)........----------------- ( -17.20, z-score = -1.23, R) >dp4.chr3 4562146 84 - 19779522 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCCACACAACUCUCCUUCCCUU------CCCCUCUCG------------------ ...(((((((........)))))))(((((....)))))((((((-(.(....).)))))))..............------.........------------------ ( -19.90, z-score = -4.58, R) >droPer1.super_2 4751022 84 - 9036312 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCCACACAACUCUCCUUCCCUU------CCCCCCUCG------------------ ...(((((((........)))))))(((((....)))))((((((-(.(....).)))))))..............------.........------------------ ( -19.90, z-score = -4.65, R) >droAna3.scaffold_13266 1090571 81 - 19884421 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCUCUGGUUCCUGCCCCCCC------CCUCCU--------------------- ...(((((((........)))))))(((((....))))).....(-(((...((.......))....)))).....------......--------------------- ( -15.30, z-score = -1.48, R) >droWil1.scaffold_180745 796563 109 + 2843958 CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUCUGUGUGUGCUGUGUGGCGCCCGCUAAAGGCAUAGGAACCAGGCAACUGGAAACCGAGAACUACUUUGA ...(((((((........)))))))..........((((((....((((((...((((....))))..))))))((..((((....))))...))))).)))....... ( -33.10, z-score = -1.62, R) >consensus CCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG_GCGCCCCCUCUCCACGUACUCCACCCCGAUAGU___CCCCCUUUGU_________________ ...(((((((........)))))))(((((....))))).(((....)))........................................................... (-10.88 = -10.89 + 0.01)
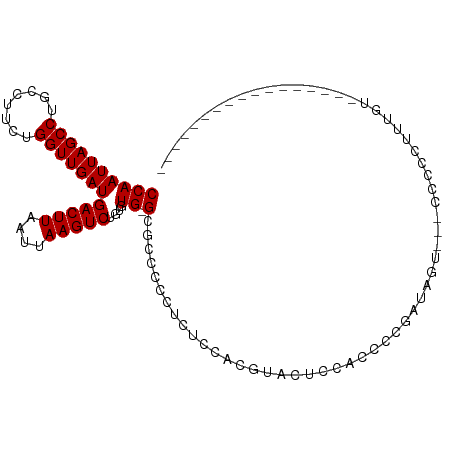
| Location | 7,379,606 – 7,379,699 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.58 |
| Shannon entropy | 0.38809 |
| G+C content | 0.54569 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7379606 93 - 21146708 ---GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCAC-GUUCUCCACCC--------------------- ---(((((....)))))(((........(((((((........)))))))(((((....))))))))(((-(.(......).))))-...........--------------------- ( -24.10, z-score = -1.02, R) >droEre2.scaffold_4845 4170669 93 + 22589142 ---GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCUUCUCCAC-GUACUCCUCCC--------------------- ---(((((....)))))(((........(((((((........)))))))(((((....))))))))(((-(.(......).))))-...........--------------------- ( -24.10, z-score = -1.12, R) >droYak2.chr2R 11555533 92 + 21139217 ---GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCUUCUCCAC-GUACUCCCCC---------------------- ---(((((....)))))(((........(((((((........)))))))(((((....))))))))(((-(.(......).))))-..........---------------------- ( -24.10, z-score = -1.17, R) >droSec1.super_1 4930907 93 - 14215200 ---GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCAC-GUACUCCACAC--------------------- ---(((((....)))))...........(((((((........)))))))(((((....))))).(((((-(.(..(.........-)..).))))))--------------------- ( -25.20, z-score = -1.14, R) >droSim1.chr2R 5902949 93 - 19596830 ---GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCCAC-GUACUCCACCC--------------------- ---(((((....)))))(((........(((((((........)))))))(((((....))))))))(((-(.(......).))))-...........--------------------- ( -24.10, z-score = -0.96, R) >dp4.chr3 4562158 94 - 19779522 ---GCUGUGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCCACACAACUCUCCUUCC--------------------- ---(((((....)))))(.(((......(((((((........)))))))(((((....)))))((((((-(.(....).)))))))..))).)....--------------------- ( -26.70, z-score = -2.40, R) >droPer1.super_2 4751034 94 - 9036312 ---GCUGUGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCCACACAACUCUCCUUCC--------------------- ---(((((....)))))(.(((......(((((((........)))))))(((((....)))))((((((-(.(....).)))))))..))).)....--------------------- ( -26.70, z-score = -2.40, R) >droAna3.scaffold_13266 1090581 96 - 19884421 GCUGCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCCCCUCUCUCUGGUUCCUGCCC---------------------- ...(((((....)))))((((....((((((((((........)))))))(((((....)))))......-..............)))...))))..---------------------- ( -26.70, z-score = -0.80, R) >droWil1.scaffold_180745 796578 116 + 2843958 ---CCUCCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUCUGUGUGUGCUGUGUGGCGCCCGCUAAAGGCAUAGGAACCAGGCAACUGGAAAC ---..(((.......(((((((......(((((((........)))))))(((((....))))))))))))(((((...((((....))))..))))).))).((((....)))).... ( -42.20, z-score = -2.43, R) >droVir3.scaffold_12875 7518011 77 + 20611582 ---GCUGUGAUCGCGGCACAGAUCUUCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCUGGCU-------------------------------------- ---((((....(((.(((((((...(((.((((((........)))))))))(((....)))))))))).-)))..)))).-------------------------------------- ( -24.70, z-score = -1.59, R) >droMoj3.scaffold_6496 6493790 77 + 26866924 ---ACUGUGAUCGCGGCACAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCUGGCU-------------------------------------- ---...((.(.(((.(((((........(((((((........)))))))(((((....)))))))))).-)))..).)).-------------------------------------- ( -23.00, z-score = -1.24, R) >droGri2.scaffold_15245 7266754 77 - 18325388 ---GCUGUGAUCGCGGCACAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG-GCGCCUGGCA-------------------------------------- ---((((....(((.(((((........(((((((........)))))))(((((....)))))))))).-)))..)))).-------------------------------------- ( -25.10, z-score = -1.42, R) >consensus ___GCUGCGAUCGCGGCGCAGAUCUCCAAUUAGCCUGCCUUCUGGUUGAUGACUUAAUUAAGUCUGUGUG_GCGCCCCCUCUCCAC_GUUCUCC_CCC_____________________ ...(((((....)))))(((........(((((((........)))))))(((((....)))))..))).................................................. (-17.85 = -17.68 + -0.18)

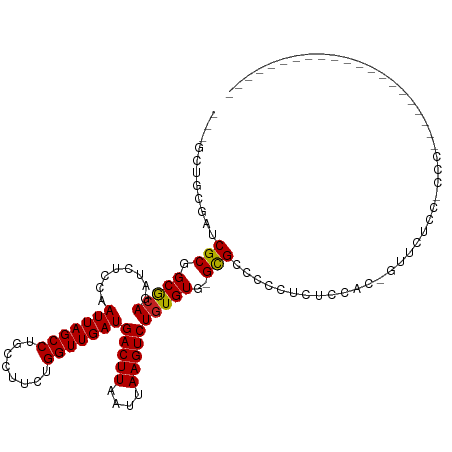
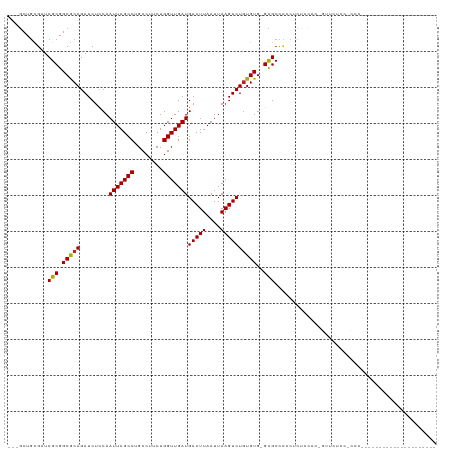
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:26 2011