| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,305,717 – 7,305,776 |
| Length | 59 |
| Max. P | 0.993339 |

| Location | 7,305,717 – 7,305,776 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 61.49 |
| Shannon entropy | 0.68834 |
| G+C content | 0.51335 |
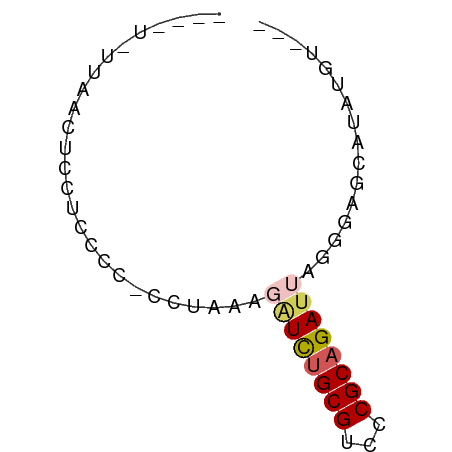
| Mean single sequence MFE | -12.68 |
| Consensus MFE | -6.66 |
| Energy contribution | -7.18 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
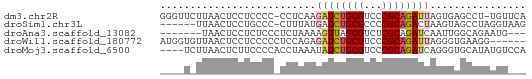
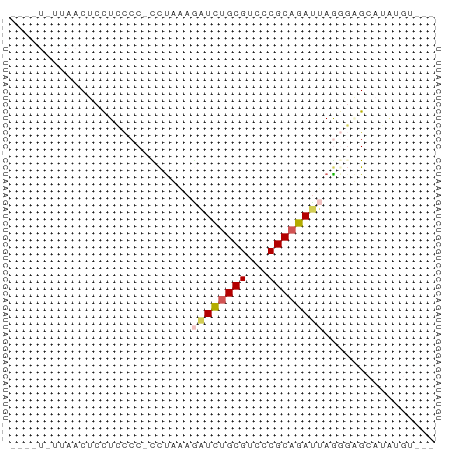
>dm3.chr2R 7305717 59 - 21146708 GGGUUCUUAACUCCUCCCC-CCUCAAGAUCUGCGUCCCGCAGAUUAGUGAGCCU-UGUUCA (((............))).-.((((.((((((((...))))))))..))))...-...... ( -15.50, z-score = -2.30, R) >droSim1.chr3L 2988948 54 + 22553184 ------UUAACUCCUGCCC-CUUUAUGAUCUGCGCCCCGCAGACUAAGUAGCCUAGGUAAG ------......(((((..-(((...(.((((((...))))))).)))..))..))).... ( -12.30, z-score = -1.81, R) >droAna3.scaffold_13082 3665230 51 - 3919855 -------UAACUCCUCUCCCUCUAAAAGUUAGCGUCUCGCAGAUCAAUUGGCAGAAUG--- -------.......(((.((.......(((.(((...))).))).....)).)))...--- ( -5.10, z-score = 0.40, R) >droWil1.scaffold_180772 3791391 55 - 8906247 AUGGUGUUAACUCCUCCCCCUCCAGAGAUCUGCGUCCCGCAGAUUAGGGUGAAGG------ ............(((.(((((.....((((((((...)))))))))))).).)))------ ( -16.70, z-score = -1.24, R) >droMoj3.scaffold_6500 646093 57 + 32352404 ----UCUUAACUCUUCCCCACCUAAAUAUCUGCGUCCCGCAGAUCAGGGUGCAUAUGUCCA ----..............(((((....(((((((...)))))))..))))).......... ( -13.80, z-score = -3.06, R) >consensus ____U_UUAACUCCUCCCC_CCUAAAGAUCUGCGUCCCGCAGAUUAGGGAGCAUAUGU___ ..........................((((((((...))))))))................ ( -6.66 = -7.18 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:17 2011