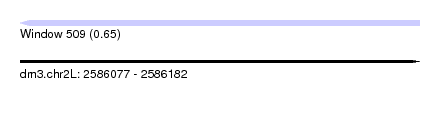
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,586,077 – 2,586,182 |
| Length | 105 |
| Max. P | 0.650243 |

| Location | 2,586,077 – 2,586,182 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.06 |
| Shannon entropy | 0.57942 |
| G+C content | 0.55001 |
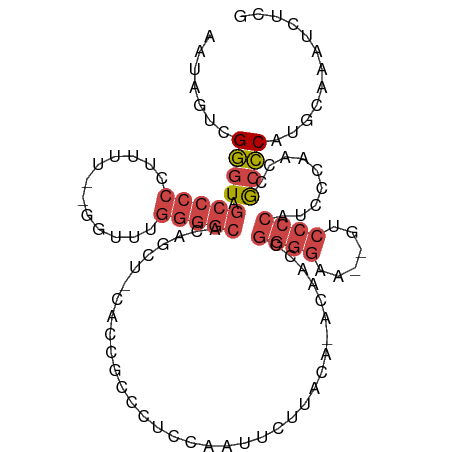
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -15.91 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
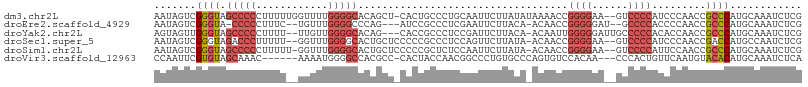
>dm3.chr2L 2586077 105 - 23011544 AAUAGUCGGGUAGCCCCCUUUUUGGUUUUGGGGCACAGCU-CACUGCCCUGCAAUUCUUAUAUAAAACCGGGGAA--GUCCCCAUCCCAACCGCCCAUGCAAAUCUCG .......((((((((........))))((((((((..((.-....))..))).................((((..--..))))..)))))..))))............ ( -28.80, z-score = -0.51, R) >droEre2.scaffold_4929 2627591 99 - 26641161 AAUAGUCGGGUA-CCCCCUUUC--UGUUUGGGGCCCAG---AUCCGCCCUCGAAUUCUUACA-ACAACCGGGGGAU--GCCCCACCCCAACCGCCCAUGCAAAUCUCG ....((.(((((-(((((....--.(((((((((....---....)))).))))).......-......))))).)--)))).)).......((....))........ ( -28.07, z-score = -0.30, R) >droYak2.chr2L 2573733 102 - 22324452 AGUAGUUGGGUAGCCCCCUUUU--UUGUUGGGGCACAG---CACCGCCCUCCGAUUCUUACA-ACAAUUGGGGGAUUGCCCCCACACCAACCGCCCAUGCAAAUCUCG .(..(((((((.(((((.....--.....)))))...(---((...(((.((((((......-..)))))))))..)))....)).)))))..).............. ( -31.70, z-score = -0.94, R) >droSec1.super_5 754338 103 - 5866729 AAUAGUCGGGUAGACCCUUUUU--GGUUUGGGGCACUGCUCCCCCGCCCUCCAGUUCUUAUA-ACAACCGGGGAA--GUCCCCAUCCCAACCGACCAUGCCAAUCUCG ....((((((....)))...((--((..(((((.(((....(((((................-.....))))).)--)))))))..))))..)))............. ( -31.80, z-score = -1.46, R) >droSim1.chr2L 2544430 104 - 22036055 AAUAGUCGGGUAGCCCCCUUUUU-GGUUUGGGGCACUGCUCCCCCGCUCUCCAAUUCUUAUA-ACAACCGGGGAA--GUCCCCAUUCCAACCGCCCAUGCAAAUCUCG .......(((..(........((-((..(((((.(((....(((((................-.....))))).)--)))))))..)))))..)))............ ( -29.30, z-score = -1.16, R) >droVir3.scaffold_12963 11153407 98 - 20206255 CCAAUUCGUGUAGCAAAC------AAAAUGGGGCCACGCC-CACUACCAACGGCCCUGUGCCCAGUGUCCACAA---CCCACUGUUCAAUGUACACAUGCAAAUCUCA .......(((((.((...------...((((((((.....-..........))))))))(..(((((.......---..)))))..)..)))))))............ ( -22.66, z-score = -0.54, R) >consensus AAUAGUCGGGUAGCCCCCUUUU__GGUUUGGGGCACAGCU_CACCGCCCUCCAAUUCUUACA_ACAACCGGGGAA__GUCCCCAUCCCAACCGCCCAUGCAAAUCUCG .......((((.(((((............)))))...................................((((......)))).........))))............ (-15.91 = -17.17 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:43 2011