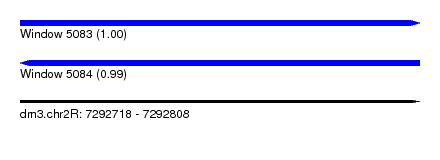
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,292,718 – 7,292,808 |
| Length | 90 |
| Max. P | 0.996061 |

| Location | 7,292,718 – 7,292,808 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.38378 |
| G+C content | 0.49653 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7292718 90 + 21146708 -------AAGAUUCAGAUUUCCAUCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.......((((....))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -27.00, z-score = -1.84, R) >droSim1.chr2R 5822742 90 + 19596830 -------AAGAUUCAGAUUUCCAUCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.......((((....))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -27.00, z-score = -1.84, R) >droSec1.super_1 4849034 90 + 14215200 -------AAUAUUCAGAUUUCCAUCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.......((((....))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -27.00, z-score = -2.30, R) >droYak2.chr2R 11471630 90 - 21139217 -------AAGAUUGAAAAUCCCUUCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.(((..(......)..)))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.50, z-score = -2.36, R) >droEre2.scaffold_4845 4088030 90 - 22589142 -------AAGAAUGGGAUUCCCAUCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.(((.((((...)))))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -31.70, z-score = -2.79, R) >droAna3.scaffold_13266 997101 90 + 19884421 -------AUUGUCACUUUUUCCAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------...................(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -2.15, R) >dp4.chr3 6301532 87 - 19779522 -------AAUGUAGAUAAUUC---UUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC -------..............---..(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -2.81, R) >droPer1.super_2 8142339 87 + 9036312 -------AAUGUAGAUAAUUC---UUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC -------..............---..(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -2.81, R) >droWil1.scaffold_180700 1999634 93 - 6630534 AGAAAUUACUUUCGGUUUGU----UUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC .((((....)))).......----..(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -27.20, z-score = -1.99, R) >droVir3.scaffold_12875 7408990 85 - 20611582 -------CAUUUCG-UAAUU----UUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------.......-.....----..(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -2.59, R) >droGri2.scaffold_15245 7154359 75 + 18325388 ------------------GA----UUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC ------------------..----..(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -3.61, R) >droMoj3.scaffold_6496 6380363 72 - 26866924 -------------------------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -------------------------.(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -3.28, R) >anoGam1.chrU 56108621 91 - 59568033 -------AAAAUUCAAAAAAGGAUCUGCCCCGAGUGAGGAUCGAACUCACGACCUUAAGAUUAUGAGACUUACGCGCUACCGACUGCGCUAUCGAGGC -------...................(((.((((((((.......)))))...((((......))))......((((........))))..))).))) ( -21.80, z-score = -1.27, R) >consensus _______AAUAUUGAUAUUUC___CUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC_ACUGAGCUAAAGAGGC ..........................(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))) (-26.31 = -25.90 + -0.41)

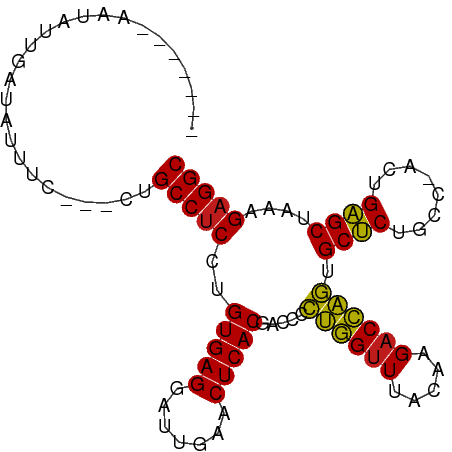
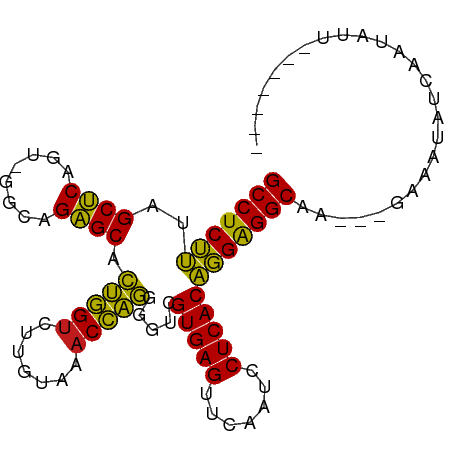
| Location | 7,292,718 – 7,292,808 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.38378 |
| G+C content | 0.49653 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -28.39 |
| Energy contribution | -27.55 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7292718 90 - 21146708 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGAUGGAAAUCUGAAUCUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))((((....)))).......------- ( -31.00, z-score = -1.60, R) >droSim1.chr2R 5822742 90 - 19596830 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGAUGGAAAUCUGAAUCUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))((((....)))).......------- ( -31.00, z-score = -1.60, R) >droSec1.super_1 4849034 90 - 14215200 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGAUGGAAAUCUGAAUAUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))((((....)))).......------- ( -31.00, z-score = -1.92, R) >droYak2.chr2R 11471630 90 + 21139217 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGAAGGGAUUUUCAAUCUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))....(((((.....)))))------- ( -28.20, z-score = -1.02, R) >droEre2.scaffold_4845 4088030 90 + 22589142 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGAUGGGAAUCCCAUUCUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).((((((...))))))...------- ( -32.90, z-score = -1.86, R) >droAna3.scaffold_13266 997101 90 - 19884421 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUGGAAAAAGUGACAAU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))...................------- ( -27.60, z-score = -1.15, R) >dp4.chr3 6301532 87 + 19779522 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA---GAAUUAUCUACAUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))..---..............------- ( -27.60, z-score = -2.05, R) >droPer1.super_2 8142339 87 - 9036312 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA---GAAUUAUCUACAUU------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))..---..............------- ( -27.60, z-score = -2.05, R) >droWil1.scaffold_180700 1999634 93 + 6630534 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA----ACAAACCGAAAGUAAUUUCU (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))..----.......((((....)))). ( -30.30, z-score = -2.63, R) >droVir3.scaffold_12875 7408990 85 + 20611582 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA----AAUUA-CGAAAUG------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))..----.....-.......------- ( -27.60, z-score = -2.09, R) >droGri2.scaffold_15245 7154359 75 - 18325388 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA----UC------------------ (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))..----..------------------ ( -27.60, z-score = -2.69, R) >droMoj3.scaffold_6496 6380363 72 + 26866924 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA------------------------- (((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).------------------------- ( -27.60, z-score = -2.19, R) >anoGam1.chrU 56108621 91 + 59568033 GCCUCGAUAGCGCAGUCGGUAGCGCGUAAGUCUCAUAAUCUUAAGGUCGUGAGUUCGAUCCUCACUCGGGGCAGAUCCUUUUUUGAAUUUU------- (((((((..((((........))))......((((..(((....)))..))))............)))))))...................------- ( -25.80, z-score = -1.17, R) >consensus GCCUCUUUAGCUCAGU_GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAA___GAAAUAUCAAUAUU_______ (((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).......................... (-28.39 = -27.55 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:15 2011