
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,292,175 – 7,292,291 |
| Length | 116 |
| Max. P | 0.992131 |

| Location | 7,292,175 – 7,292,291 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.57429 |
| G+C content | 0.50597 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -29.47 |
| Energy contribution | -27.13 |
| Covariance contribution | -2.34 |
| Combinations/Pair | 1.86 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7292175 116 + 21146708 UUACUCUUUUGGCGACCGCCUUGCUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUAUGAAAACCCGAA-- ...(((((.....((((....(((((..(((.((.......)).))--).)))))(((((.......))))).))))(((((.......))))))))))...................-- ( -34.10, z-score = -0.37, R) >droSim1.chr2R 5822218 116 + 19596830 UUACUCUUUUGCCAGCCGCCUUGCUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUAUGAAAACCUACU-- ....((..(((((..(((((((((((..(((.((.......)).))--).)))))(((((.......))))))))).(((((.......))))).)).)))))....)).........-- ( -34.70, z-score = -0.82, R) >droSec1.super_1 4848507 116 + 14215200 UUACUUAUUUGCCAGCCGCCUUGCUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUAUGAAAACCUACU-- ...(.((.(((((..(((((((((((..(((.((.......)).))--).)))))(((((.......))))))))).(((((.......))))).)).))))).)).)..........-- ( -34.80, z-score = -1.04, R) >droYak2.chr2R 11471193 114 - 21139217 UUUCUCUUUUGCCGGCCGCCUUGCCCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAG--UUAAAGGCCUACU-- ....(((((((((((((((.((((((.(((.......)))..).))--))).)).(((((.......))))).))))(((((.......)))))....)))).--..)))))......-- ( -36.40, z-score = -0.15, R) >droEre2.scaffold_4845 4087507 116 - 22589142 UUUCUCUUUUGCCGGCCGCCUUGCCCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGCUUUUAAAACCUACU-- ........(((((((((((.((((((.(((.......)))..).))--))).)).(((((.......))))).))))(((((.......)))))....)))))...............-- ( -35.90, z-score = -0.56, R) >droAna3.scaffold_13266 996589 109 + 19884421 UUUCGCUUUUGCCGGCCGCGUUGCUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUUCGUAAAU--------- ....((...((((((((....(((((..(((.((.......)).))--).)))))(((((.......))))).))))(((((.......)))))....))))...))....--------- ( -36.30, z-score = -1.43, R) >dp4.chr3 6301169 116 - 19779522 UACUGCUUUUGCCACGCGCGUCACUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUACGAAAAAUUUAU-- ...((((((((((..((..((((((..(((.......))).)))))--)...)).(((((.......))))).))).(((((.......))))).)))))))................-- ( -32.20, z-score = -0.74, R) >droPer1.super_2 8141977 116 + 9036312 UACUGCUUUUGCCACGCGCGUCACUCAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUAUGAAAAAUUUAU-- ...((((((((((..((..((((((..(((.......))).)))))--)...)).(((((.......))))).))).(((((.......))))).)))))))................-- ( -32.20, z-score = -0.66, R) >droWil1.scaffold_181009 3119985 102 - 3585778 UAGGCAUUUUGACCAUCAUCACACACAUGGGGAUGUAGCUCAGUGG--UAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCAUCUCCAGU---------------- ..(((.....).)).............((((((((...((((...(--((((((....)))))))...)))).....(((((.......)))))))))))))..---------------- ( -34.10, z-score = -0.73, R) >droVir3.scaffold_12875 7408496 115 - 20611582 AUCGAUUUUAGCGAGGCGCGUUGCUCAUGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUCUUCAAAUAGUU--- ...((((((.(((((((.((.(((((.((((.((.......)).))--))))))).)))))))))......))))))(((((.......))))).(((((....)))))........--- ( -34.20, z-score = -0.90, R) >droMoj3.scaffold_6496 6379498 115 - 26866924 AUAGGUUUUAGCGUGGCGCGUUGCUCAUGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAACUCCGAAACUAGAU--- ...(((((..(((.(((.((.(((((.((((.((.......)).))--))))))).))))).)))))))).((..(((((((.......))))).((((....))))))..))....--- ( -43.50, z-score = -2.66, R) >droGri2.scaffold_15245 7153648 117 + 18325388 AUCGGUUUUAGCGGGGCGCGUUGCUCAUGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAA-CUUCGAAAAUUACAC ...(((((..(((((((.((.(((((.((((.((.......)).))--))))))).)))))))))))))).((((...((....))..))))...((((.....-))))........... ( -38.70, z-score = -1.76, R) >anoGam1.chr2R 48739919 106 + 62725911 UUGUGCUUUUAGCGACUUGAAUGCACACGGCUCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCAAUGA-------------- .(((((.(((((....))))).))))).(((((((((((...))))...((((.((((.(((....((((....).))).))).)))).)))).))))))).....-------------- ( -38.10, z-score = -1.92, R) >consensus UUACGCUUUUGCCGGCCGCCUUGCUCAAGCCUCUUUAGCUCAGUGG__CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAUUAUGAAAACCUAAU__ ............................(((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))................... (-29.47 = -27.13 + -2.34)



| Location | 7,292,175 – 7,292,291 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.57429 |
| G+C content | 0.50597 |
| Mean single sequence MFE | -35.71 |
| Consensus MFE | -27.04 |
| Energy contribution | -25.78 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7292175 116 - 21146708 --UUCGGGUUUUCAUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGCAAGGCGGUCGCCAAAAGAGUAA --(((.(((.................(((((.......)))))(((((((((((.....))))..(((((.(--((.((.......)).)))..)))))))).)))))))....)))... ( -37.50, z-score = -1.31, R) >droSim1.chr2R 5822218 116 - 19596830 --AGUAGGUUUUCAUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGCAAGGCGGCUGGCAAAAGAGUAA --...............(((((.((.(((((.......)))))(.((.((((((.....))))))(((((.(--((.((.......)).)))..))))).)))))..)))))........ ( -37.40, z-score = -1.02, R) >droSec1.super_1 4848507 116 - 14215200 --AGUAGGUUUUCAUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGCAAGGCGGCUGGCAAAUAAGUAA --...............(((((.((.(((((.......)))))(.((.((((((.....))))))(((((.(--((.((.......)).)))..))))).)))))..)))))........ ( -37.40, z-score = -1.29, R) >droYak2.chr2R 11471193 114 + 21139217 --AGUAGGCCUUUAA--CUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGGGCAAGGCGGCCGGCAAAAGAGAAA --.((.((((.....--........((((((.......))))))....((((((.....)))))).((..((--((...(((((.....))))).))))..)))))).)).......... ( -40.00, z-score = -1.10, R) >droEre2.scaffold_4845 4087507 116 + 22589142 --AGUAGGUUUUAAAAGCUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGGGCAAGGCGGCCGGCAAAAGAGAAA --..............((((((.((((((((.......))))).....((((((.....))))))(((((.(--((.((.......)).)))..)))))))).)).)))).......... ( -38.80, z-score = -0.77, R) >droAna3.scaffold_13266 996589 109 - 19884421 ---------AUUUACGAAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGCAACGCGGCCGGCAAAAGCGAAA ---------.....((..((((....(((((.......)))))(.((.((((((.....))))))(((((.(--((.((.......)).)))..)))))....)).)))))....))... ( -35.60, z-score = -2.25, R) >dp4.chr3 6301169 116 + 19779522 --AUAAAUUUUUCGUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGUGACGCGCGUGGCAAAAGCAGUA --..............(((((.....(((((.......)))))((.(.((((((.....)))))).).))((--((((.(((((.....)))))..(((...)))))))))...))))). ( -34.00, z-score = -1.26, R) >droPer1.super_2 8141977 116 - 9036312 --AUAAAUUUUUCAUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUGAGUGACGCGCGUGGCAAAAGCAGUA --..............(((((.....(((((.......)))))((.(.((((((.....)))))).).))((--((((.(((((.....)))))..(((...)))))))))...))))). ( -34.00, z-score = -1.47, R) >droWil1.scaffold_181009 3119985 102 + 3585778 ----------------ACUGGAGAUGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUA--CCACUGAGCUACAUCCCCAUGUGUGUGAUGAUGGUCAAAAUGCCUA ----------------...((.....(((((.......)))))(((((((((((((((..(.((..((((..--.....))))....)).)..))))))))).)).))))......)).. ( -35.40, z-score = -1.43, R) >droVir3.scaffold_12875 7408496 115 + 20611582 ---AACUAUUUGAAGAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCAUGAGCAACGCGCCUCGCUAAAAUCGAU ---.....((((..((..(((....((((((.......))))))....((((((.....))))))(((((((--((.((.......)).)))).)))))..)))..))..))))...... ( -32.30, z-score = -2.26, R) >droMoj3.scaffold_6496 6379498 115 + 26866924 ---AUCUAGUUUCGGAGUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCAUGAGCAACGCGCCACGCUAAAACCUAU ---.....(..((((((((.((((....))))....))))).)))..)((((((.....))))))(((((((--((.((.......)).)))).)))))..(((...))).......... ( -35.70, z-score = -2.05, R) >droGri2.scaffold_15245 7153648 117 - 18325388 GUGUAAUUUUCGAAG-UUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCAUGAGCAACGCGCCCCGCUAAAACCGAU ((((.....(((.((-(((.((((....))))...)))))..)))...((((((.....))))))(((((((--((.((.......)).)))).)))))))))................. ( -34.30, z-score = -2.24, R) >anoGam1.chr2R 48739919 106 - 62725911 --------------UCAUUGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCGUUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGAGCCGUGUGCAUUCAAGUCGCUAAAAGCACAA --------------.....(((((((..((((.(((......))).))))((.......)).....((((.........))))..))))))).(((((................))))). ( -31.79, z-score = -1.28, R) >consensus __AGUAGGUUUUCAUAAUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG__CCACUGAGCUAAAGAGGCUUGAGCAACGCGGCCGGCAAAAGCGGAA ...................(((((..(((((.......))))).....((((((.....)))))).((((.........))))....)))))............................ (-27.04 = -25.78 + -1.27)

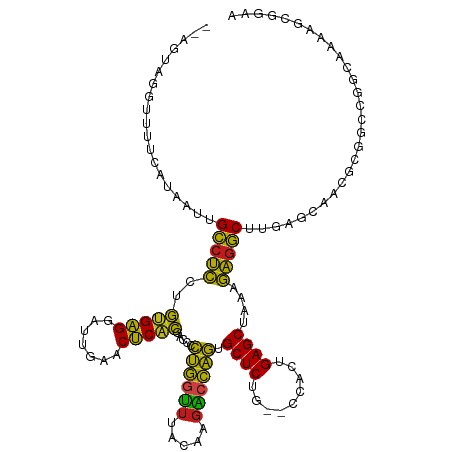
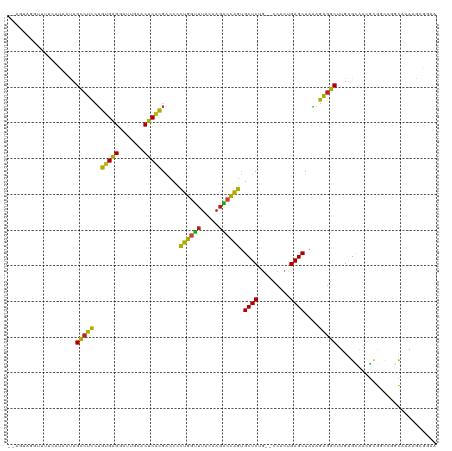
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:13 2011