
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,287,274 – 7,287,394 |
| Length | 120 |
| Max. P | 0.986900 |

| Location | 7,287,274 – 7,287,394 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 73.65 |
| Shannon entropy | 0.57389 |
| G+C content | 0.54618 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.50 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
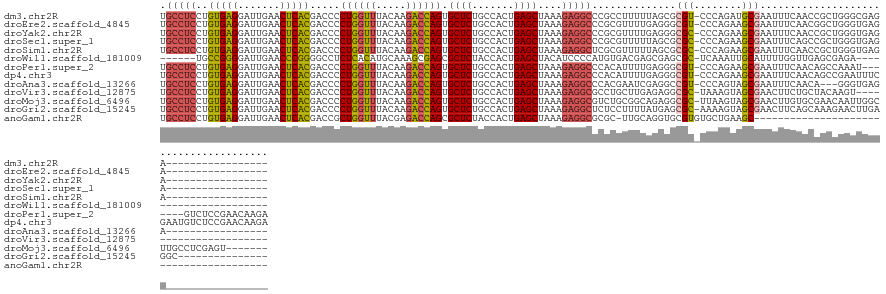
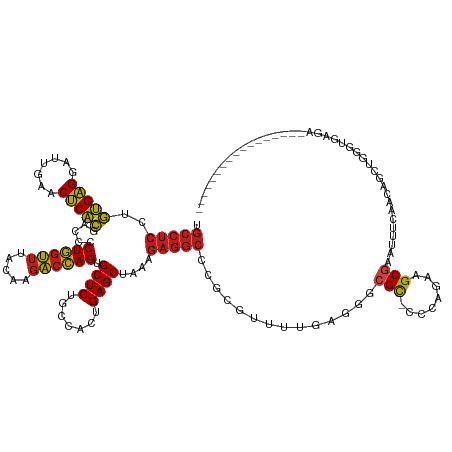
>dm3.chr2R 7287274 120 + 21146708 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCGCCUUUUUAGCGCGU-CCCAGAUGCGAAUUUCAACCGCUGGGCGAGA----------------- ...(((.((((((.......))))))....((((((.....)))))).((((.((...(((((((((((((....))))))))))((((-(...))))).....)))...)).))))))).----------------- ( -44.90, z-score = -3.35, R) >droEre2.scaffold_4845 4082635 120 - 22589142 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCGCGUUUUGAGGGCGU-CCCAGAAGCGAAUUUCAACGGCUGGGUGAGA----------------- ..((((....))))......(((((.....((((((.....)))))).((((.......))))....((.((((.........)))).)-)((((...((........)).))))))))).----------------- ( -38.70, z-score = -0.26, R) >droYak2.chr2R 11466276 120 - 21139217 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCGCGUUUUGAGGGCGC-CCCAGAAGCGAAUUUCAACCGCUGGGUGAGA----------------- .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....))))).(((.(((((.((....-))))))))))..(((((.((....)))))))----------------- ( -41.60, z-score = -1.49, R) >droSec1.super_1 4843675 120 + 14215200 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCGCGUUUUUAGCGCGC-CCCAGAAGCGAAUUUCAGCCGCUGGGUGAGA----------------- .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....))))).(((((.....))))).-(((((..((........))..))))).....----------------- ( -44.60, z-score = -2.61, R) >droSim1.chr2R 5816770 120 + 19596830 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCUCGCGUUUUUAGCGCGC-CCCAGAAGCGAAUUUCAACCGCUGGGUGAGA----------------- .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....)))))..((((.....))))((-((....((((.........))))))))....----------------- ( -41.40, z-score = -1.89, R) >droWil1.scaffold_181009 3119574 109 - 3585778 ------UGCCGGGGAUUGAACCCGGGGCCUCUCACAUGCAAAGCGAGCGCUCUACCACUGAGCUACAUCCCCAUGUGACGAGCGAGCGC-UCAAAUUGCAUUUUGGUUGAGCGAGA---------------------- ------..(((((.......)))))....((((....((.......))((((.((((..(.(((((((....)))))..((((....))-)).....)).)..)))).))))))))---------------------- ( -36.20, z-score = -0.62, R) >droPer1.super_2 8134838 130 + 9036312 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCACAUUUUGAGGGCGU-CCCAGAAGCGAAUUUCAACAGCCAAAU-------GUCUCCGAACAAGA .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....)))))......((((..(((((-.......((...........))...))-------)))..))))..... ( -33.20, z-score = 0.02, R) >dp4.chr3 6294013 137 - 19779522 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCACAUUUUGAGGGCGU-CCCAGAAGCGAAUUUCAACAGCCGAAUUUCGAAUGUCUCCGAACAAGA .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....))))).....(((((.(((...-)))(((..((((.(((.......))).))))....))).....))))) ( -38.00, z-score = -0.87, R) >droAna3.scaffold_13266 991833 117 + 19884421 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCACGAAUCGAGGCCGU-CCCAGUAGCGAAUUUCAACA---GGGUGAGA----------------- ...(((.((((((.......))))))..(((((...........(((((......))))).((((..(((((((.......).)))).)-)....)))).........))---))).))).----------------- ( -35.20, z-score = -0.60, R) >droVir3.scaffold_12875 7402392 115 - 20611582 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCGCCUGCUUGAGAGGCGC-UAAAGUAGCGAACUUCUGCUACAAGU---------------------- .(((((.((((((.......))))))....((((((.....)))))).((((.......))))....)))))((((.......))))..-....((((((......))))))....---------------------- ( -41.10, z-score = -2.47, R) >droMoj3.scaffold_6496 6370905 130 - 26866924 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCGUCUGCGGCAGAGGCGC-UUAAGUAGCGAACUUGUGCGAACAAUUGGCUUGCCUCGAGU------- ((((....(((((.......)))))(((((((((((.....)))))).((((.......))))......)).)))...))))(((((((-....))(((.((.((((....)))))).))).)))))....------- ( -45.20, z-score = -1.09, R) >droGri2.scaffold_15245 7147148 122 + 18325388 UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCUCUCCUUUUAUGAGCGC-AAAAGUAGCGAACUUCAGCAAAGAACUUGAGGC--------------- .(((((.((((((.......))))))....((((((.....)))))).((((.......(((((.....))))).........))))((-..((((.....))))..)).........)))))--------------- ( -36.39, z-score = -1.15, R) >anoGam1.chr2R 25327851 98 + 62725911 UGCCUCCUGUGAGGAUUGAACUCACGACCGCUGGUUUACGAGACCAGCGCUCUACCACUGAGCUAAAGAGGCGCGC-UUGCAGGUGCGUGUGCUGAAGC--------------------------------------- .(((((.((((((.......))))))...(((((((.....)))))))((((.......))))....)))))((((-.(((....))).))))......--------------------------------------- ( -41.60, z-score = -3.01, R) >consensus UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCCACUGAGCUAAAGAGGCCCGCGUUUUGAGGGCGC_CCCAGAAGCGAAUUUCAACAGCUGGGUGAGA_________________ .(((((..(((((.......))))).....((((((.....)))))).((((.......))))....)))))..............(((........)))...................................... (-26.76 = -26.50 + -0.26)
| Location | 7,287,274 – 7,287,394 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 138 |
| Reading direction | reverse |
| Mean pairwise identity | 73.65 |
| Shannon entropy | 0.57389 |
| G+C content | 0.54618 |
| Mean single sequence MFE | -42.41 |
| Consensus MFE | -27.38 |
| Energy contribution | -26.77 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
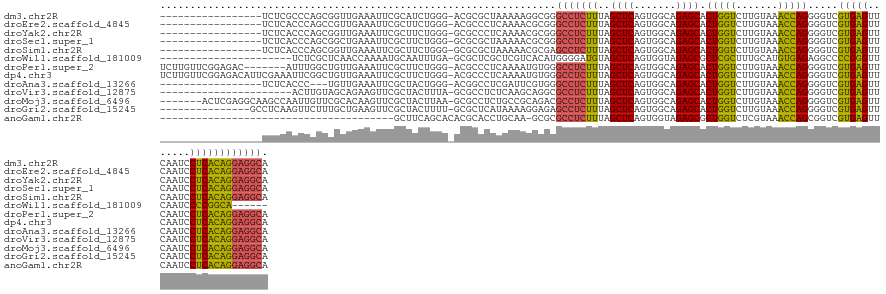
>dm3.chr2R 7287274 120 - 21146708 -----------------UCUCGCCCAGCGGUUGAAAUUCGCAUCUGGG-ACGCGCUAAAAAGGCGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------((((((((.((((.......))))..((((.-((..((((((.(((....))).))))))((((((......))))))....))...)))))))).(((((.......)))))..)))).. ( -43.30, z-score = -1.02, R) >droEre2.scaffold_4845 4082635 120 + 22589142 -----------------UCUCACCCAGCCGUUGAAAUUCGCUUCUGGG-ACGCCCUCAAAACGCGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------((((((((.((((((((.....(((...(((-(.((((.(.....).)))).)))).)))))))))))......(((((.......))))))))).(((((.......)))))..)))).. ( -42.50, z-score = -1.18, R) >droYak2.chr2R 11466276 120 + 21139217 -----------------UCUCACCCAGCGGUUGAAAUUCGCUUCUGGG-GCGCCCUCAAAACGCGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------.....(((((.(((........))).)))))-..(((..(........(((((.....((((.......)))).(((((.......))))))))))(((((.......))))).)..))). ( -41.70, z-score = -0.54, R) >droSec1.super_1 4843675 120 - 14215200 -----------------UCUCACCCAGCGGCUGAAAUUCGCUUCUGGG-GCGCGCUAAAAACGCGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------((((((((.((.(((((.....(((...(((-((.(((.......))).)))))...)))))))).))......(((((.......))))))))).(((((.......)))))..)))).. ( -49.00, z-score = -2.51, R) >droSim1.chr2R 5816770 120 - 19596830 -----------------UCUCACCCAGCGGUUGAAAUUCGCUUCUGGG-GCGCGCUAAAAACGCGAGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------.....(((((.(((........))).)))))-.((((.......)))).(((((((..((((.......)))).(((((.......))))).....(((((.......)))))))))))). ( -43.10, z-score = -1.25, R) >droWil1.scaffold_181009 3119574 109 + 3585778 ----------------------UCUCGCUCAACCAAAAUGCAAUUUGA-GCGCUCGCUCGUCACAUGGGGAUGUAGCUCAGUGGUAGAGCGCUCGCUUUGCAUGUGAGAGGCCCCGGGUUCAAUCCCCGGCA------ ----------------------((((((.........((((((..(((-((((((.....((((.((((.......))))))))..)))))))))..)))))))))))).(((..(((......))).))).------ ( -44.20, z-score = -2.32, R) >droPer1.super_2 8134838 130 - 9036312 UCUUGUUCGGAGAC-------AUUUGGCUGUUGAAAUUCGCUUCUGGG-ACGCCCUCAAAAUGUGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA (((((((((....)-------.....((..((((.....(((...(((-(.((((.(.....).)))).)))).)))))))..)).)))).(((((.......))))).....(((((.......))))))))).... ( -40.90, z-score = 0.20, R) >dp4.chr3 6294013 137 + 19779522 UCUUGUUCGGAGACAUUCGAAAUUCGGCUGUUGAAAUUCGCUUCUGGG-ACGCCCUCAAAAUGUGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA ((((((..(((((((..((.....))..))))(((...((((((((((-(.((((.(.....).)))).))...((..(((((......)))))..))......))))))).))....)))..)))..)))))).... ( -44.30, z-score = -0.47, R) >droAna3.scaffold_13266 991833 117 - 19884421 -----------------UCUCACCC---UGUUGAAAUUCGCUACUGGG-ACGGCCUCGAUUCGUGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -----------------((((((((---...........(((((((((-..(((((........))))).......)))))))))......(((((.......))))))))).(((((.......)))))..)))).. ( -41.30, z-score = -0.80, R) >droVir3.scaffold_12875 7402392 115 + 20611582 ----------------------ACUUGUAGCAGAAGUUCGCUACUUUA-GCGCCUCUCAAGCAGGCGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA ----------------------....(((((........)))))....-..(((((....).))))(((((((..((((.......)))).(((((.......))))).....(((((.......)))))))))))). ( -39.50, z-score = -1.01, R) >droMoj3.scaffold_6496 6370905 130 + 26866924 -------ACUCGAGGCAAGCCAAUUGUUCGCACAAGUUCGCUACUUAA-GCGCCUCUGCCGCAGACGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA -------.((..((...(((...((((....))))....))).))..)-).(((((((((((.((.((.......)))).))))).((.(.(((((.......))))).).))(((((.......))))).)))))). ( -42.20, z-score = -0.31, R) >droGri2.scaffold_15245 7147148 122 - 18325388 ---------------GCCUCAAGUUCUUUGCUGAAGUUCGCUACUUUU-GCGCUCAUAAAAGGAGAGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA ---------------(((((..(((((..(((((((...(((.(((((-(......))))))...)))..)))))))..)).))).((.(.(((((.......))))).).))(((((.......)))))..))))). ( -41.90, z-score = -1.33, R) >anoGam1.chr2R 25327851 98 - 62725911 ---------------------------------------GCUUCAGCACACGCACCUGCAA-GCGCGCCUCUUUAGCUCAGUGGUAGAGCGCUGGUCUCGUAAACCAGCGGUCGUGAGUUCAAUCCUCACAGGAGGCA ---------------------------------------(((((((.........))).))-))..(((((((.............((.(((((((.......))))))).))(((((.......)))))))))))). ( -37.40, z-score = -2.03, R) >consensus _________________UCUCACCCAGCUGUUGAAAUUCGCUUCUGGG_GCGCCCUCAAAACGCGGGCCUCUUUAGCUCAGUGGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA ..................................................................(((((((..((((.......)))).(((((.......))))).....(((((.......)))))))))))). (-27.38 = -26.77 + -0.61)
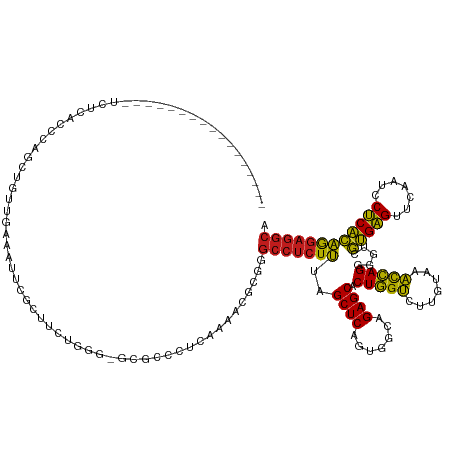
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:12 2011