| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,281,823 – 7,281,929 |
| Length | 106 |
| Max. P | 0.989286 |

| Location | 7,281,823 – 7,281,929 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Shannon entropy | 0.48106 |
| G+C content | 0.52056 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -27.89 |
| Energy contribution | -27.05 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
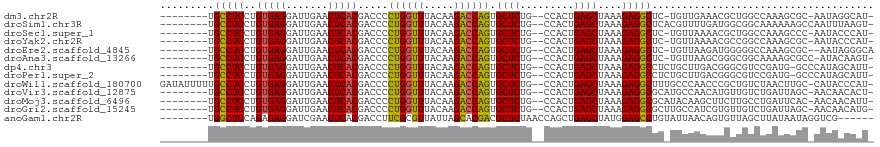
>dm3.chr2R 7281823 106 + 21146708 -AUGCCUAUU-GCGCUUUGGCCAGCGUUUCAACA-GAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.(((((...-..((((((.(((..((....)).-((((.......))))..)))--)))))).(((((.......))))).....(((((.......)))))...)))))-------- ( -34.20, z-score = -0.32, R) >droSim1.chr3R 14158387 108 - 27517382 -ACUUAAAUUGGCUUUUUUGCCGCCAUCAAAACGUGAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.........(((......)))(((.((.......((.((......((((.....--..)))).(((((.......))))))).))(((((.......)))))..))))).-------- ( -33.60, z-score = -1.00, R) >droSec1.super_1 4838290 106 + 14215200 -AUGGGUAUU-GGGCUUUGGCCAGCGUUUUAACA-GAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.((((.(((-(((((((((((...(((((..((-((((.......))))..)).--.))))).(((((.......))))).))))).))))))))).)))).........-------- ( -33.90, z-score = -0.27, R) >droYak2.chr2R 11460899 106 - 21139217 -AUGGGUAUU-GCGCUUUGGCCGGCGUUUUAACA-GAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.((..((..-(((((......)))))..)).))-..(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -34.90, z-score = -0.45, R) >droEre2.scaffold_4845 4076162 106 - 22589142 UGCCCUAUU--GCGCUUUGGCCCCCAUCUUAACA-GAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- .((......--))(((((((((((.......(((-(..((......((((.....--..))))...))..))))......))))))(((((.......)))))..))))).-------- ( -33.12, z-score = 0.00, R) >droAna3.scaffold_13266 986439 106 + 19884421 -ACUUGUAU-GGCGCUUUUGCCGCCCGCUUAACA-GAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -........-(((((....).)))).........-..(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -33.70, z-score = 0.06, R) >dp4.chr3 6287788 107 - 19779522 -AAUGCUAUGGGC-CAUCGGACGCCCGUCAAGCAGAGGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -..((((((((((-........))))))..))))...(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -40.10, z-score = -1.39, R) >droPer1.super_2 8128453 107 + 9036312 -AAUGCUAUGGGC-CAUCGGACGCCCGUCAAGCAGAGGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -..((((((((((-........))))))..))))...(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -40.10, z-score = -1.39, R) >droWil1.scaffold_180700 1982260 115 - 6630534 -AUGGGUAUG-GCAAGUUAGACAGCGGGUUGGGCAAAGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAAAAUAUC -...(((((.-....((..(((.....)))..))...(((((((..((((.....--..)))).(((((.......))))).....(((((.......))))))))))))....))))) ( -32.00, z-score = -0.10, R) >droVir3.scaffold_12875 7391762 107 - 20611582 -AGUGUUGUU-GCUAAUCAGACAACAUGUUGGCAUGCGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.((((((((-........))))))))..........(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -35.80, z-score = -0.95, R) >droMoj3.scaffold_6496 6364058 107 - 26866924 -AAUGUUGUU-GUGAAUCAGGCAAGAAGCUUGUAUGCGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -.........-(((...(((((.....)))))..)))(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -33.60, z-score = -0.56, R) >droGri2.scaffold_15245 7138650 107 + 18325388 -CAUGUUGUU-GCUAAUCAGACAACACGAUGGCAAGCGCCUCUUUAGCUCAGUGG--CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA-------- -..(((((((-........)))))))...........(((((((..((((.....--..)))).(((((.......))))).....(((((.......)))))))))))).-------- ( -34.20, z-score = -0.88, R) >anoGam1.chr2R 13947255 105 - 62725911 ------CGACCUAUUAUAAGCUAACACUGUUAAUACAGGCUCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCA-------- ------........(((.(((.......))).)))..(((((((((((...))))...((((.((((.(((....((((....).))).))).)))).)))).))))))).-------- ( -30.80, z-score = -1.03, R) >consensus _AAUGGUAUU_GCGCAUUGGCCACCAUUUUAACA_GAGCCUCUUUAGCUCAGUGG__CAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCA________ .....................................(((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))......... (-27.89 = -27.05 + -0.84)

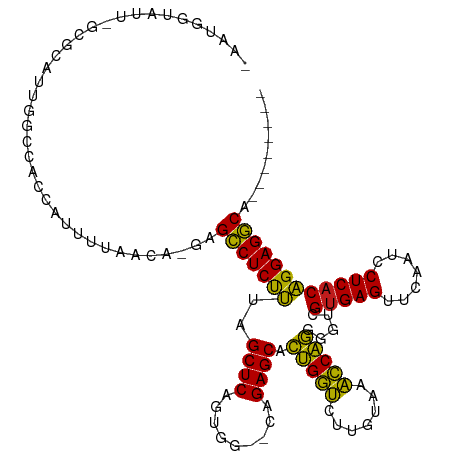
| Location | 7,281,823 – 7,281,929 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.08 |
| Shannon entropy | 0.48106 |
| G+C content | 0.52056 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -26.82 |
| Energy contribution | -26.41 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7281823 106 - 21146708 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUC-UGUUGAAACGCUGGCCAAAGCGC-AAUAGGCAU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))((-(((((...((((......)))))-))))))...- ( -37.40, z-score = -2.48, R) >droSim1.chr3R 14158387 108 + 27517382 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUCACGUUUUGAUGGCGGCAAAAAAGCCAAUUUAAGU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))...........((((.........))))........- ( -32.80, z-score = -1.60, R) >droSec1.super_1 4838290 106 - 14215200 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUC-UGUUAAAACGCUGGCCAAAGCCC-AAUACCCAU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))..-............(((....))).-.........- ( -30.50, z-score = -2.03, R) >droYak2.chr2R 11460899 106 + 21139217 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUC-UGUUAAAACGCCGGCCAAAGCGC-AAUACCCAU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))..-........(((........))).-.........- ( -29.70, z-score = -1.45, R) >droEre2.scaffold_4845 4076162 106 + 22589142 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUC-UGUUAAGAUGGGGGCCAAAGCGC--AAUAGGGCA --------.((((((((((((.......))))))....((((((.....))))))...(((.--..((.(((((.....)))))-.))..))).))))))....((.(--....).)). ( -36.30, z-score = -0.78, R) >droAna3.scaffold_13266 986439 106 - 19884421 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUC-UGUUAAGCGGGCGGCAAAAGCGCC-AUACAAGU- --------((((....(((((.......)))))(.(((((((((.....)))))).(((..(--(....(((((.....)))))-.))..)))))))))))........-........- ( -35.10, z-score = -1.18, R) >dp4.chr3 6287788 107 + 19779522 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCCUCUGCUUGACGGGCGUCCGAUG-GCCCAUAGCAUU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))...((((....((((........-))))..))))..- ( -38.00, z-score = -2.12, R) >droPer1.super_2 8128453 107 - 9036312 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCCUCUGCUUGACGGGCGUCCGAUG-GCCCAUAGCAUU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))...((((....((((........-))))..))))..- ( -38.00, z-score = -2.12, R) >droWil1.scaffold_180700 1982260 115 + 6630534 GAUAUUUUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCUUUGCCCAACCCGCUGUCUAACUUGC-CAUACCCAU- ((((.....(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))...((.......))))))........-.........- ( -28.80, z-score = -2.09, R) >droVir3.scaffold_12875 7391762 107 + 20611582 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCGCAUGCCAACAUGUUGUCUGAUUAGC-AACAACACU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....)))))...........((((((.......))-)))).....- ( -32.80, z-score = -2.11, R) >droMoj3.scaffold_6496 6364058 107 + 26866924 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCGCAUACAAGCUUCUUGCCUGAUUCAC-AACAACAUU- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....))))).....((.((.....)).))......-.........- ( -30.50, z-score = -2.15, R) >droGri2.scaffold_15245 7138650 107 - 18325388 --------UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG--CCACUGAGCUAAAGAGGCGCUUGCCAUCGUGUUGUCUGAUUAGC-AACAACAUG- --------.(((((.((((((.......))))))....((((((.....)))))).((((..--.....))))....))))).........((((((((.((.....)-)))))))))- ( -35.40, z-score = -2.65, R) >anoGam1.chr2R 13947255 105 + 62725911 --------UGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCGUUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGAGCCUGUAUUAACAGUGUUAGCUUAUAAUAGGUCG------ --------.(((((((..((((.(((......))).))))((.......)).....((((.........))))..)))))))(.(((((..(((....)))..))))).)...------ ( -28.10, z-score = -0.36, R) >consensus ________UGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUG__CCACUGAGCUAAAGAGGCUC_UGUUAAAAUGGUGGCCAAAGCGC_AAUAACAAU_ .........(((((..(((((.......))))).....((((((.....)))))).((((.........))))....)))))..................................... (-26.82 = -26.41 + -0.41)
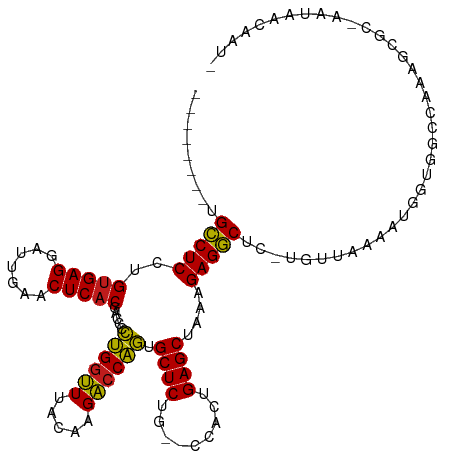
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:10 2011