| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,260,942 – 7,261,043 |
| Length | 101 |
| Max. P | 0.915449 |

| Location | 7,260,942 – 7,261,043 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.52732 |
| G+C content | 0.50049 |
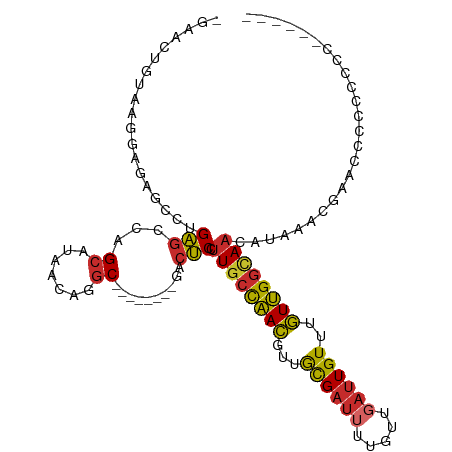
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -12.05 |
| Energy contribution | -11.74 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
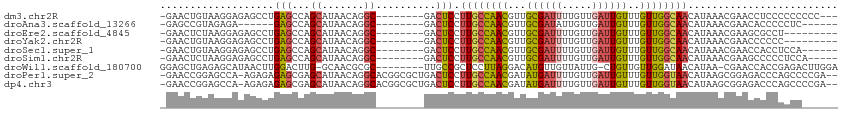
>dm3.chr2R 7260942 101 + 21146708 -GAACUGUAAGGAGAGCCUGAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAACCUCCCCCCCCC--- -....(((((((((.(((((...........)))))--------..))))))(((((((..((((((.....)))))).))))))).)))....................--- ( -30.90, z-score = -3.52, R) >droAna3.scaffold_13266 958647 92 + 19884421 -GAGCCGUAGAGA------GAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUAUUGUUGAUUGUUUGUUGGCAACAUAAACGAACACCCCCUC------ -....(((...((------(.(((.........)))--------..))).(((((((((..(((((.......))))).))))))))).....)))...........------ ( -22.70, z-score = -1.35, R) >droEre2.scaffold_4845 4055331 95 - 22589142 -GAACUCUAAGGAGAGCCUGAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAAGCGCCU--------- -.......((((((.(((((...........)))))--------..))))))(((((((..((((((.....)))))).)))))))..................--------- ( -30.50, z-score = -2.39, R) >droYak2.chr2R 11439702 95 - 21139217 -GAACUGUAAGGAGAGCCUGAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAACCCCCC--------- -....(((((((((.(((((...........)))))--------..))))))(((((((..((((((.....)))))).))))))).)))..............--------- ( -30.90, z-score = -3.63, R) >droSec1.super_1 4817587 98 + 14215200 -GAACUGUAAGGAGAGCCUGAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAACCACCUCCA------ -....(((((((((.(((((...........)))))--------..))))))(((((((..((((((.....)))))).))))))).))).................------ ( -30.90, z-score = -2.88, R) >droSim1.chr2R 5795123 99 + 19596830 -GAACUCUAAGGAGAGCCUGAGCCAGCAUAACAGGC--------GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAAGCCCCCUCCA----- -.......((((((.(((((...........)))))--------..))))))(((((((..((((((.....)))))).)))))))......................----- ( -30.50, z-score = -2.64, R) >droWil1.scaffold_180700 1943558 102 - 6630534 GGAGCUGAGAGCAUAACUUGGACUUG-GCAACGCGC--------UUGCCGCUCCUUAGGACAUGUUGUUAUUG-CUGUUGUUGGAUAACAUAA-CGAACCACCGAGACUUGGA (((((.(((..((.....))..)))(-((((.....--------))))))))))(((((..((((((((...(-(....))..))))))))..-((......))...))))). ( -24.80, z-score = -0.12, R) >droPer1.super_2 8104187 109 + 9036312 -GAACCGGAGCCA-AGAGAGAGCGAGCAUAACAGGCACGGCGCUGACUCCUUGCCAACGAUAUGAUUUUGUUGAUUGUUUGUUGGUAACAUAAGCGGAGACCCAGCCCCGA-- -........(((.-...................))).(((.((((.((((((((((((((.(..(((.....)))..))))))))))).......))))...)))).))).-- ( -30.06, z-score = -1.29, R) >dp4.chr3 6263474 109 - 19779522 -GAACCGGAGCCA-AGAGAGAGCGAGCAUAACAGGCACGGCGCUGACUCCUUGCCAACGAUAUGAUUUUGUUGAUUGUUUGUUGGUAACAUAAGCGGAGACCCAGCCCCGA-- -........(((.-...................))).(((.((((.((((((((((((((.(..(((.....)))..))))))))))).......))))...)))).))).-- ( -30.06, z-score = -1.29, R) >consensus _GAACUGUAAGGAGAGCCUGAGCCAGCAUAACAGGC________GACUCCUUGCCAACGUUGCGAUUUUGUUGAUUGUUUGUUGGCAACAUAAACGAACCCCCCCCC______ ..................................................((((((((...((((((.....))))))..))))))))......................... (-12.05 = -11.74 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:06 2011