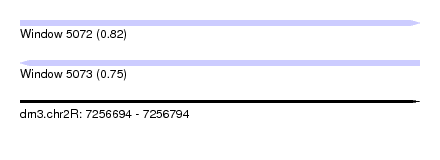
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,256,694 – 7,256,794 |
| Length | 100 |
| Max. P | 0.819795 |

| Location | 7,256,694 – 7,256,794 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.17 |
| Shannon entropy | 0.69556 |
| G+C content | 0.64172 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
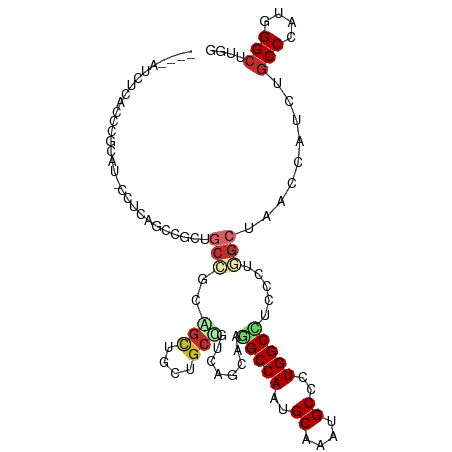
>dm3.chr2R 7256694 100 + 21146708 ----AUCUCACCCGCAU-CCUCAGUCGCUGCCGCAGCUGCUGCUGUCAGCAAGGCCAAUGCAAAUGCCCUGGCCUCUCUGGCUAACCAUCUGCCCAUGGGCUUGG ----.............-....((((((((..(((((....))))))))).((((((..((....))..))))))....))))..(((...(((....))).))) ( -36.20, z-score = -1.39, R) >droSim1.chr2R 5790826 100 + 19596830 ----AUCUCACCCGCAU-CCUCAGCCGCUGCCGCAGCUGCUGCUGUCAGCAAGGCCAAUGCAAAUGCGCUGGCCUCUCUGGCUAACCAUCUGCCCAUGGGCUUGG ----.............-....((((((((..(((((....))))))))).((((((.(((....))).))))))....))))..(((...(((....))).))) ( -40.50, z-score = -1.98, R) >droSec1.super_1 4813307 100 + 14215200 ----AUCUCACCCGCAU-CCUCAGCCGCUGCCGCAGCUGCUGCUGUCAGCAAGGCCAAUGCAAAUGCCCUGGCCUCACUGGCUAACCAUCUGCCCAUGGGCUUGG ----.............-....((((((((..(((((....))))))))).((((((..((....))..))))))....))))..(((...(((....))).))) ( -38.90, z-score = -1.78, R) >droYak2.chr2R 11435596 100 - 21139217 ----AUCUCGCCCGCAU-CCUCAGCCGCUGCCGCAGCUGCUGCCGUCAGCAAGGCCAAUGCAAAUGCACUGGCCUCUUUGGCUAACCAUCUGCCCAUGGGCUUGG ----.............-....(((((((((.((((...)))).).)))).((((((.(((....))).))))))....))))..(((...(((....))).))) ( -39.10, z-score = -1.53, R) >droEre2.scaffold_4845 4051642 100 - 22589142 ----AUCUCACCCGCAU-CCUCGGCCGCUGCCGCAGCUGCUGCCGUCAGCAAGGCCAAUGCAAAUGCACUGGCCUCACUGGCUAACCAUCUGCCCAUGGGCUUGG ----.............-....(((.((.((....)).)).)))(((((..((((((.(((....))).))))))..)))))...(((...(((....))).))) ( -40.50, z-score = -2.08, R) >droAna3.scaffold_13266 954937 88 + 19884421 ----------------G-UCUCACCUGCGACAUCAGCUGCGGCUGUCUCGAAGGCCAAUGCUAAUGCCCUGGCUUCCUUAGCCACACACCUGCCCUUGGGAAUGG ----------------(-((.(....).)))..((((....))))((((((.(((....((....))..(((((.....))))).......))).)))))).... ( -24.80, z-score = 0.10, R) >droVir3.scaffold_12875 7357940 100 - 20611582 GCC-UUGCCGCUCA-GUGCCGCUGGCAAGGCUUCGGUGGCGGCCUCGGUUAGUGCCAACGCCGCGGCGCUGGCGACGCUAGCUA---GUCUGCCCAUGGGCAUUG (((-(((((((...-.....)).))))))))....((.(((((...(((....)))...))))).))((((((...))))))..---...((((....))))... ( -48.30, z-score = -0.33, R) >droGri2.scaffold_15245 7102507 100 + 18325388 GCUGCCGCCGCUGCCAUGCCGCU--CAAGGCAUCUGUGGCGGCGUCGGUGAGCGCCAAUGCCGCUGCUCUGGCCACCUUGGCCA---GUCUGCCCAUGGGCAUUG (((((((.((((((((((((...--...))))....)))))))).)))).)))..(((((((.(.((.((((((.....)))))---)...))....)))))))) ( -53.90, z-score = -2.67, R) >droMoj3.scaffold_6496 6323390 88 - 26866924 GCC-CUGCCCCUGA-GUGCGGCGGGCAAGAG------------CUCAGUCAGUGCCAAUGCCGCUGCGCUGGCCACCCUAGCCA---GCCUGCCCAUGGGCAUCG ...-.(((((.((.-(((((((((.((...(------------(.(.....).))...)))))))))((((((.......))))---))..)).)).)))))... ( -38.10, z-score = -0.26, R) >consensus ____AUCUCACCCGCAU_CCUCAGCCGCUGCCGCAGCUGCUGCCGUCAGCAAGGCCAAUGCAAAUGCCCUGGCCUCCCUGGCUAACCAUCUGCCCAUGGGCUUGG .............................(((..(((....)))........(((((..((....))..))))).....))).........(((....))).... (-15.75 = -16.43 + 0.69)


| Location | 7,256,694 – 7,256,794 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.17 |
| Shannon entropy | 0.69556 |
| G+C content | 0.64172 |
| Mean single sequence MFE | -44.19 |
| Consensus MFE | -15.25 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7256694 100 - 21146708 CCAAGCCCAUGGGCAGAUGGUUAGCCAGAGAGGCCAGGGCAUUUGCAUUGGCCUUGCUGACAGCAGCAGCUGCGGCAGCGACUGAGG-AUGCGGGUGAGAU---- ((..(((....))).....((((((....((((((((.((....)).)))))))))))))).(((.(((.(((....))).)))...-.))))).......---- ( -39.80, z-score = -0.81, R) >droSim1.chr2R 5790826 100 - 19596830 CCAAGCCCAUGGGCAGAUGGUUAGCCAGAGAGGCCAGCGCAUUUGCAUUGGCCUUGCUGACAGCAGCAGCUGCGGCAGCGGCUGAGG-AUGCGGGUGAGAU---- ((..(((....))).....((((((....((((((((.((....)).)))))))))))))).(((.(((((((....)))))))...-.))))).......---- ( -46.40, z-score = -2.08, R) >droSec1.super_1 4813307 100 - 14215200 CCAAGCCCAUGGGCAGAUGGUUAGCCAGUGAGGCCAGGGCAUUUGCAUUGGCCUUGCUGACAGCAGCAGCUGCGGCAGCGGCUGAGG-AUGCGGGUGAGAU---- (((.(((....)))...)))(((.((((..(((((((.((....)).)))))))..))....(((.(((((((....)))))))...-.))))).)))...---- ( -47.60, z-score = -2.24, R) >droYak2.chr2R 11435596 100 + 21139217 CCAAGCCCAUGGGCAGAUGGUUAGCCAAAGAGGCCAGUGCAUUUGCAUUGGCCUUGCUGACGGCAGCAGCUGCGGCAGCGGCUGAGG-AUGCGGGCGAGAU---- ....((((...........((((((....(((((((((((....))))))))))))))))).(((.(((((((....)))))))...-.))))))).....---- ( -52.60, z-score = -3.75, R) >droEre2.scaffold_4845 4051642 100 + 22589142 CCAAGCCCAUGGGCAGAUGGUUAGCCAGUGAGGCCAGUGCAUUUGCAUUGGCCUUGCUGACGGCAGCAGCUGCGGCAGCGGCCGAGG-AUGCGGGUGAGAU---- ((..(((....))).........(((((..((((((((((....))))))))))..))).((((.((.((....)).)).))))...-..)))).......---- ( -49.00, z-score = -2.57, R) >droAna3.scaffold_13266 954937 88 - 19884421 CCAUUCCCAAGGGCAGGUGUGUGGCUAAGGAAGCCAGGGCAUUAGCAUUGGCCUUCGAGACAGCCGCAGCUGAUGUCGCAGGUGAGA-C---------------- ......((((..((..((((.(((((.....))))).).)))..)).))))(((.(((.(((((....)))).).))).))).....-.---------------- ( -29.10, z-score = 0.01, R) >droVir3.scaffold_12875 7357940 100 + 20611582 CAAUGCCCAUGGGCAGAC---UAGCUAGCGUCGCCAGCGCCGCGGCGUUGGCACUAACCGAGGCCGCCACCGAAGCCUUGCCAGCGGCAC-UGAGCGGCAA-GGC ...((((....))))...---..(((.((...)).)))...(((((.((((......)))).))))).......((((((((.((.....-...)))))))-))) ( -44.30, z-score = -0.52, R) >droGri2.scaffold_15245 7102507 100 - 18325388 CAAUGCCCAUGGGCAGAC---UGGCCAAGGUGGCCAGAGCAGCGGCAUUGGCGCUCACCGACGCCGCCACAGAUGCCUUG--AGCGGCAUGGCAGCGGCGGCAGC ...((((....))))..(---(((((.....)))))).((.((.((...((((........))))((((....((((...--...)))))))).)).)).))... ( -46.40, z-score = -0.81, R) >droMoj3.scaffold_6496 6323390 88 + 26866924 CGAUGCCCAUGGGCAGGC---UGGCUAGGGUGGCCAGCGCAGCGGCAUUGGCACUGACUGAG------------CUCUUGCCCGCCGCAC-UCAGGGGCAG-GGC ...(((((.((((...((---(((((.....)))))))((.((((((..(((.(.....).)------------))..)).)))).)).)-))).))))).-... ( -42.50, z-score = -0.28, R) >consensus CCAAGCCCAUGGGCAGAUGGUUAGCCAGAGAGGCCAGCGCAUUUGCAUUGGCCUUGCUGACAGCAGCAGCUGCGGCAGCGGCUGAGG_AUGCGGGUGAGAU____ ....(((....))).........(((......(((((.((....)).))))).........((......))..)))............................. (-15.25 = -15.53 + 0.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:05 2011