| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,246,448 – 7,246,516 |
| Length | 68 |
| Max. P | 0.866807 |

| Location | 7,246,448 – 7,246,516 |
|---|---|
| Length | 68 |
| Sequences | 13 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.47328 |
| G+C content | 0.49599 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.55 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866807 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
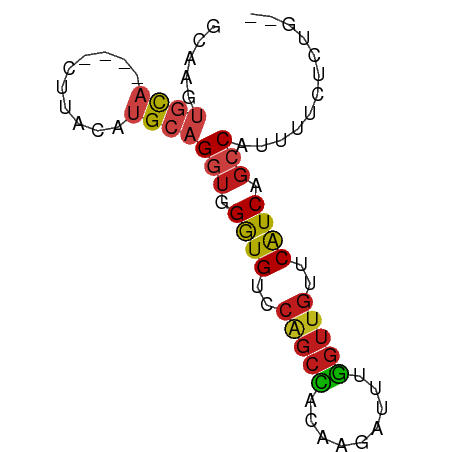
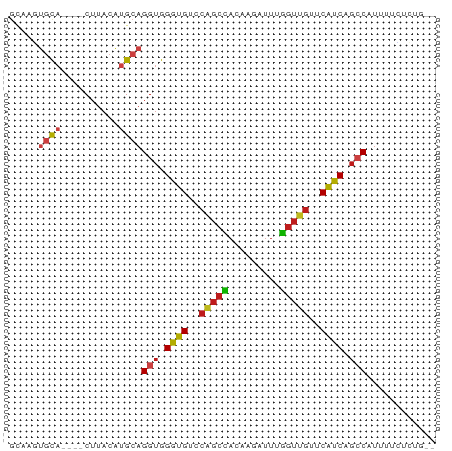
>dm3.chr2R 7246448 68 + 21146708 GCAAGUGCA----CUUACAUGCAGGUGGGUGUCCGGCCACAAGAUUUGGUUGUUCAUCUGCCAUUUGUUCUG-- (((((((((----......))).((..((((..((((((.......))))))..))))..))))))))....-- ( -24.30, z-score = -2.15, R) >droEre2.scaffold_4845 4039912 68 - 22589142 GCAAGUGCA----CUUACAUGCAGGUGGGUGUCCGGCCACAAGAUUUGGUUGUUCAUCUGCCAUUUUCUCUG-- (((.(((..----..))).))).((..((((..((((((.......))))))..))))..))..........-- ( -23.30, z-score = -2.17, R) >droYak2.chr2R 11424072 68 - 21139217 GAAAGUGCA----CUUACAUGCAGGUGGGUGUCCGGCUACAAGAUUUGGUUGUUCAUCCGCCAUUUUCUCCG-- (((((((((----......))).((((((((..((((((.......))))))..))))))))))))))....-- ( -25.10, z-score = -3.33, R) >droSec1.super_1 4802778 68 + 14215200 GCAAGUGCA----CUUACAUGCAGGUGGGUGUCCGGCCACAAGAUUUGGUUGUUCAUCUGCCAUUUUCUCUG-- (((.(((..----..))).))).((..((((..((((((.......))))))..))))..))..........-- ( -23.30, z-score = -2.17, R) >droSim1.chr2R_random 2014587 68 + 2996586 GCAAGUGCA----CUUACAUGCAGGUGGGUGUCCGGCCACAAGAUUUGGUUGUUCAUCUGCCAUUUCCUCUG-- (((.(((..----..))).))).((..((((..((((((.......))))))..))))..))..........-- ( -23.30, z-score = -1.93, R) >dp4.chr3 14099907 68 + 19779522 GCAAUUACA----CUUACAAGCAGGUGGGUGUCCAGCCACGAGAUUGGGUUGUUCAUCAGACAUUUUCAGUA-- .......((----(((......)))))((((..(((((.((....)))))))..))))..............-- ( -15.20, z-score = -0.60, R) >droPer1.super_4 5944442 68 - 7162766 GCAAUUACA----CUUACAAGCAGGUGGGUGUCCAGCCACGAGAUUGGGUUGUUCAUCAGACAUUUUCAGUA-- .......((----(((......)))))((((..(((((.((....)))))))..))))..............-- ( -15.20, z-score = -0.60, R) >droAna3.scaffold_13266 944458 68 + 19884421 GCAAUUGCA----CUUACGUGCAGGUGGGUGUCCGGCCACCAGAUUUGGUUGCUCAUCAGCCAUUUUAUGUG-- (((..((((----(....)))))(((((.(....).))))).((..((((((.....))))))..)).))).-- ( -22.60, z-score = -1.38, R) >droWil1.scaffold_180699 2250391 64 + 2593675 GGACUUGCA----CUUACGUGCAGGUGGGUGGCCCGCCACAAGAUUUGGUUGUUCAUCAGCCAUAUUG------ ..(((((((----(....))))))))..((((....))))..(((.((((((.....)))))).))).------ ( -25.10, z-score = -2.61, R) >droGri2.scaffold_15112 680858 63 + 5172618 -----AGCA----CUUACAUGCAGGUGGGUGUCCAGCCACCAGAUUGGGUUGUUCAUCAGCCAUUGUUAGUA-- -----...(----((.(((....(((((.(....).)))))......(((((.....)))))..))).))).-- ( -18.50, z-score = -0.83, R) >droVir3.scaffold_12875 5745914 70 + 20611582 GUUAGUGCA----CUUACAUGCAGGUGGAUGUCCAGCUACCAAAUUUGGUUGUUCAUCAGCCAUGUUUACGAUC .....((((----......))))(((.((((..((((((.......))))))..)))).)))............ ( -18.30, z-score = -1.18, R) >droMoj3.scaffold_6496 2548372 70 - 26866924 CUAAAUGCA----CUUACAUGCAGGUGGGUGUCCAGCUACUAGAUUCGGUUGUUCGUCCGCCAUGUUUUAAACA .....((((----......))))(((((..(..(((((.........)))))..)..)))))............ ( -19.60, z-score = -1.63, R) >anoGam1.chr3L 8069961 72 - 41284009 GCAAAUGUAGCGCCUUACCUGCCGGGGGGUGUCCAGCGACCAGUCCUUUGUCUUCAUCGGCCAUCGUUUCUG-- .........(((((((.((....)))))))))..(((((...(.((..((....))..)))..)))))....-- ( -13.70, z-score = 2.11, R) >consensus GCAAGUGCA____CUUACAUGCAGGUGGGUGUCCAGCCACAAGAUUUGGUUGUUCAUCAGCCAUUUUCUCUG__ .......................(((.((((..(((((.........)))))..)))).)))............ (-12.67 = -12.55 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:04 2011