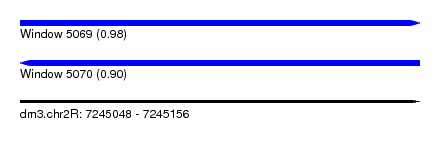
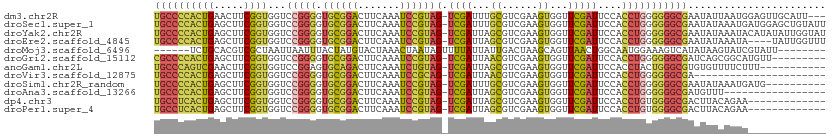
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,245,048 – 7,245,156 |
| Length | 108 |
| Max. P | 0.980771 |

| Location | 7,245,048 – 7,245,156 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.57375 |
| G+C content | 0.53226 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -20.12 |
| Energy contribution | -18.98 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
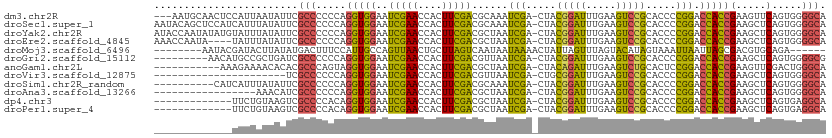
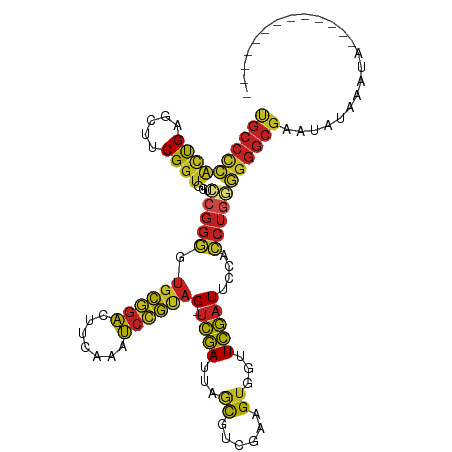
>dm3.chr2R 7245048 108 + 21146708 ---AAUGCAACUCCAUUAAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCAAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGUUCAGUGGGGCA ---.....................(((((.(.(((........)))((((((...((((((..-.....))))))..(((((.....)))))...))))))...).))))). ( -32.80, z-score = -1.69, R) >droSec1.super_1 4801404 111 + 14215200 AAUACAGCUCCAUCAUUUAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCAAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA ......(((((((........((((((.....)))))).........(((((...((((((..-.....))))))..(((((.....)))))...)))))....))))))). ( -33.30, z-score = -1.88, R) >droYak2.chr2R 11422733 111 - 21139217 AUACCAAUAUAUGUAUUUAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA .....((((((......)))))).(((((.(.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).).))))). ( -31.90, z-score = -1.50, R) >droEre2.scaffold_4845 4038416 107 - 22589142 AAACCAAUA----UAUUUAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA .........----...........(((((.(.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).).))))). ( -30.60, z-score = -1.60, R) >droMoj3.scaffold_6496 24341414 98 + 26866924 --------AAUACGAUACUUAUAUGACUUUCCAUUGCCAGUUAACUGCUUAGUCAAUAAUAAAACUAUUAGUUUAGUACAUAGUAAAUUAAUUAGCGACGUGCAGA------ --------...............(((((..........))))).((((...(((..((((.((..(((((((.....)).)))))..)).))))..)))..)))).------ ( -13.20, z-score = 0.47, R) >droGri2.scaffold_15112 679864 102 + 5172618 ---------AACAUGCCGCUGAUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGUUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCG ---------..............((((((.(.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).).)))))) ( -31.10, z-score = -0.48, R) >anoGam1.chr2L 45492148 100 + 48795086 -----------AAAGAAAACACACGCCCAGUAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACAGAUUUGAAGUCUGCACUCCGGACCACCGAAGUUCGACUGGGCA -----------.............(((((((.(((((..(((((....)))))......(((.-...(((((.....))))).....))).)))))(.....).))))))). ( -30.30, z-score = -2.31, R) >droVir3.scaffold_12875 5744900 89 + 20611582 ----------------------UCGCCCCCCAGGUGGAAUCGAACCACUUCGACGUUAAUCGA-CUGCGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA ----------------------..(((((.(.(((((..(((((....)))))......(((.-.(((((((.....)))))))...))).)))))(.....).).))))). ( -34.90, z-score = -2.33, R) >droSim1.chr2R_random 2013239 101 + 2996586 ----------CAUCAUUUAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCAAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA ----------..............(((((.(.(((........))).(((((...((((((..-.....))))))..(((((.....)))))...)))))....).))))). ( -31.60, z-score = -1.77, R) >droAna3.scaffold_13266 943309 94 + 19884421 -----------------AAACAUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA -----------------.......(((((.(.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).).))))). ( -30.60, z-score = -1.50, R) >dp4.chr3 14098677 98 + 19779522 -------------UUCUGUAAGUCGCCCCACAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGAGGCA -------------...........(((.(((.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).))).))). ( -28.50, z-score = -0.79, R) >droPer1.super_4 5943212 98 - 7162766 -------------UUCUGUAAGUCGCCCCACAGGUGGAAUCGAACCACUUCGACGCUAAUCGA-CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGAGGCA -------------...........(((.(((.(((((..(((((((...((((......))))-....)).)))))..((((.....)))))))))(.....).))).))). ( -28.50, z-score = -0.79, R) >consensus _____________UAUUUAUAUUCGCCCCCCAGGUGGAAUCGAACCACUUCGACGCUAAUCGA_CUACGGAUUUGAAGUCCGCACCCCGGACCACCGAAGCUCAGUGGGGCA ........................(((.(.(.(((((..(((((....)))))......(((.....(((((.....))))).....))).)))))(.....).).).))). (-20.12 = -18.98 + -1.13)
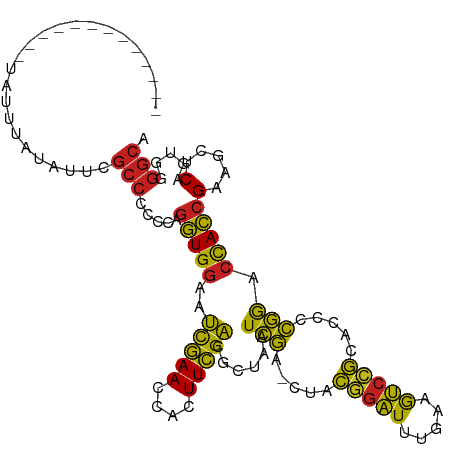
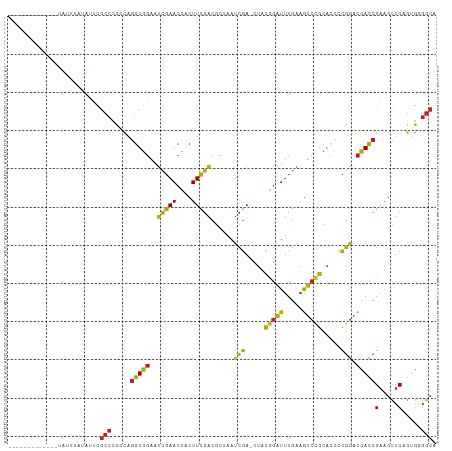
| Location | 7,245,048 – 7,245,156 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.57375 |
| G+C content | 0.53226 |
| Mean single sequence MFE | -34.31 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7245048 108 - 21146708 UGCCCCACUGAACUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUUGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUUAAUGGAGUUGCAUU--- .(((((((((.....))))...(((((.((((((.......))))))(-((((.(..(......)..).)))))....)))))))))).....................--- ( -37.30, z-score = -0.58, R) >droSec1.super_1 4801404 111 - 14215200 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUUGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUAAAUGAUGGAGCUGUAUU .(((((((((.....))))...(((((.((((((.......))))))(-((((.(..(......)..).)))))....))))))))))........................ ( -37.30, z-score = -0.25, R) >droYak2.chr2R 11422733 111 + 21139217 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUAAAUACAUAUAUUGGUAU .(((((((((.....))))...(((((.((((((.......))))))(-((((...((......))...)))))....)))))))))).((((((......))))))..... ( -37.20, z-score = -0.82, R) >droEre2.scaffold_4845 4038416 107 + 22589142 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUAAAUA----UAUUGGUUU .(((((((((.....))))...(((((.((((((.......))))))(-((((...((......))...)))))....))))))))))...........----......... ( -36.00, z-score = -0.65, R) >droMoj3.scaffold_6496 24341414 98 - 26866924 ------UCUGCACGUCGCUAAUUAAUUUACUAUGUACUAAACUAAUAGUUUUAUUAUUGACUAAGCAGUUAACUGGCAAUGGAAAGUCAUAUAAGUAUCGUAUU-------- ------.((((..((((.((((......(((((...........)))))...)))).))))...)))).....((((........))))...............-------- ( -13.40, z-score = 0.90, R) >droGri2.scaffold_15112 679864 102 - 5172618 CGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAACGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAUCAGCGGCAUGUU--------- ((((((((((.....))))...(((((.((((((.......))))))(-((((...((......))...)))))....)))))))))))..............--------- ( -37.50, z-score = -0.24, R) >anoGam1.chr2L 45492148 100 - 48795086 UGCCCAGUCGAACUUCGGUGGUCCGGAGUGCAGACUUCAAAUCUGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUACUGGGCGUGUGUUUUCUUU----------- .((((((((..(((((((....)))))))...))).........((((-(((((....))))).((((.......))))))))))))).............----------- ( -34.40, z-score = -2.09, R) >droVir3.scaffold_12875 5744900 89 - 20611582 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGCAG-UCGAUUAACGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGA---------------------- .(((((((((.....))))...(((((.((((((.......))))))(-((((...((......))...)))))....))))))))))..---------------------- ( -37.70, z-score = -1.39, R) >droSim1.chr2R_random 2013239 101 - 2996586 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUUGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUAAAUGAUG---------- .(((((((((.....))))...(((((.((((((.......))))))(-((((.(..(......)..).)))))....))))))))))..............---------- ( -37.30, z-score = -1.25, R) >droAna3.scaffold_13266 943309 94 - 19884421 UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAUGUUU----------------- .(((((((((.....))))...(((((.((((((.......))))))(-((((...((......))...)))))....)))))))))).......----------------- ( -36.00, z-score = -0.80, R) >dp4.chr3 14098677 98 - 19779522 UGCCUCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGUGGGGCGACUUACAGAA------------- .(((((((...(((((((....)))))))(((((.......)))))..-(((((....))))).((((.......))))..)))))))...........------------- ( -33.80, z-score = -0.40, R) >droPer1.super_4 5943212 98 + 7162766 UGCCUCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG-UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGUGGGGCGACUUACAGAA------------- .(((((((...(((((((....)))))))(((((.......)))))..-(((((....))))).((((.......))))..)))))))...........------------- ( -33.80, z-score = -0.40, R) >consensus UGCCCCACUGAGCUUCGGUGGUCCGGGGUGCGGACUUCAAAUCCGUAG_UCGAUUAGCGUCGAAGUGGUUCGAUUCCACCUGGGGGGCGAAUAUAAAUA_____________ ((((((((((.....))))...(((((.((((((.......))))))..((((...((......))...)))).....)))))))))))....................... (-26.86 = -26.88 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:15:03 2011