
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,147,656 – 7,147,752 |
| Length | 96 |
| Max. P | 0.812231 |

| Location | 7,147,656 – 7,147,752 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.30 |
| Shannon entropy | 0.68049 |
| G+C content | 0.51189 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -9.10 |
| Energy contribution | -8.31 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7147656 96 + 21146708 ACAUUGGGAUCGGGGCGAUGACGCCGGCGAUUGAUGACACCGAUUAUCAUUCGGCGGCCACAUUAACGGCCUCUGCA----ACUGCAUAACUUACAUGGC-------------------- .(((((((...(((((((((..((((.(((.((((((......)))))).))).))))..))))....)))))(((.----...)))...)))).)))..-------------------- ( -32.00, z-score = -2.40, R) >droSim1.chr2R 5698790 96 + 19596830 ACAUUGGGAUCGGGGCGAUGACGCCGGCGAUUGAUGACACCGAUUAUCAUUCGGCGGCCACAUUAACGGCCUCUGCA----ACUGCAUAACUUACACGGC-------------------- ....((((...(((((((((..((((.(((.((((((......)))))).))).))))..))))....)))))(((.----...)))...))))......-------------------- ( -30.10, z-score = -1.86, R) >droSec1.super_1 4716185 98 + 14215200 ACAUUGGGAUCGGGGCGAUGACGCCGGCGAUUGAUGACACAGAUUAUCAUUCGGCGGCUACAUUAACGGCCUCUGCA----ACUGCAUAACAUACACUGCAU------------------ .((.((.....(((((((((..((((.(((.((((((......)))))).))).))))..))))....)))))(((.----...))).......)).))...------------------ ( -30.10, z-score = -2.63, R) >droYak2.chr2R 19147109 75 - 21139217 ---------------------ACAUGGCGAUCGAUGACACCGAUUAUCAUUCGGCGGCCACAUUAACGGCCUCUGCA----ACUGCAUAACUUACACGGC-------------------- ---------------------....(.(((..(((((......)))))..))).)((((........))))..(((.----...))).............-------------------- ( -17.10, z-score = -1.02, R) >droEre2.scaffold_4845 3940311 84 - 22589142 ------------ACAUUGGCAUGGCGGCGAUUGAUGACACCGAUUAUCAUUCGCCGGCCACAUUAACGGCCUCUGCA----ACUGCAUAACUUACACGGC-------------------- ------------......(((..(((((((.((((((......)))))).)))))((((........))))...)).----..)))..............-------------------- ( -28.10, z-score = -3.36, R) >droAna3.scaffold_13266 856807 78 + 19884421 -----------AGUCGAAAGUUAGUUGGGAUUGAUGACACCCAUCA-------GUGCAUCCAUUAACAGCCUCUGCA----ACUGCAUAAUUCACAUAGA-------------------- -----------.((.(((.((((((.((((((((((.....)))))-------))...)))))))))......(((.----...)))...))))).....-------------------- ( -16.40, z-score = -0.56, R) >dp4.chr3 11034308 93 + 19779522 UGAGUCAGAUCUGGGUGAUGACGCCGGCGAUUGAUGACACCGAUCA-------GGGGCUCCAUUAGCGGCCUCUGCAGCUGACUGCAUAACUUACACGGC-------------------- ...(((((((((.((((....)))).).))))..)))).(((...(-------((((((.(....).)))))))((((....))))..........))).-------------------- ( -29.80, z-score = -0.52, R) >droPer1.super_4 6357090 93 + 7162766 UGAGUCAGAUCUGGGUGAUGACGCCGGCGAUUGAUGACACCGAUCA-------GGGGCUCCAUUAGCGGCCUCUGCAGCUGACUGCAUAACUUACACGGC-------------------- ...(((((((((.((((....)))).).))))..)))).(((...(-------((((((.(....).)))))))((((....))))..........))).-------------------- ( -29.80, z-score = -0.52, R) >droGri2.scaffold_15245 3037638 111 - 18325388 AUAUUUGUAUCUGUGAAUGUAUCUUUCGGAUUGAUGGCAUCAGUCA-------GCGCAUCUAUUAGCUGGCUCAAUG--CUAACAAUUAUCUUUCCAAGCAGUGAAUUAUUCGCUGCUGC (((((..(....)..)))))........(((((.((((((.(((((-------((..........)))))))..)))--))).))))).........((((((((.....)))))))).. ( -33.50, z-score = -2.66, R) >consensus A_AUU_GGAUCGGGGCGAUGACGCCGGCGAUUGAUGACACCGAUUAUCAUUCGGCGGCUACAUUAACGGCCUCUGCA____ACUGCAUAACUUACACGGC____________________ ............................(((((.......)))))..........((((........))))..((((......))))................................. ( -9.10 = -8.31 + -0.78)

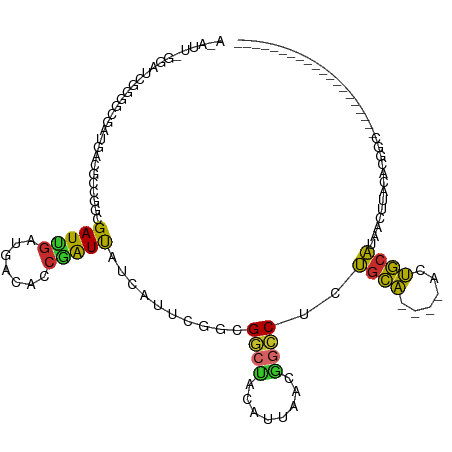
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:55 2011