| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,130,572 – 7,130,712 |
| Length | 140 |
| Max. P | 0.947527 |

| Location | 7,130,572 – 7,130,712 |
|---|---|
| Length | 140 |
| Sequences | 3 |
| Columns | 155 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Shannon entropy | 0.21328 |
| G+C content | 0.35789 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -27.05 |
| Energy contribution | -30.50 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
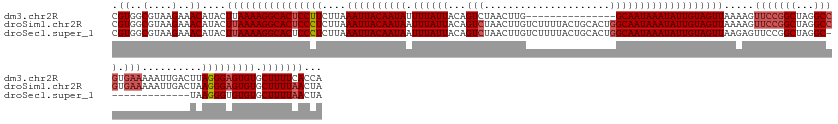
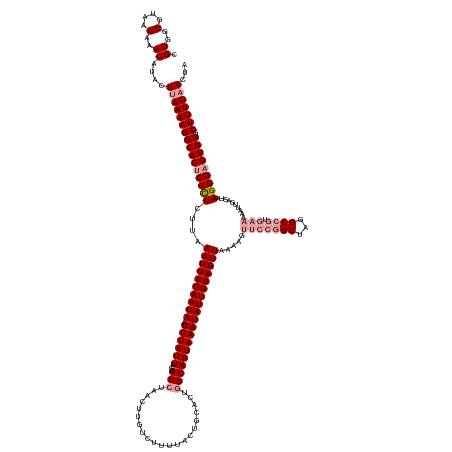
>dm3.chr2R 7130572 140 + 21146708 CGUGGCGUAAGAAACAUACUUAAAAGGCACUCCUUCUUAAAUUACAAUAUUUUAUUACAGUCUAACUUG---------------GCAAUAAAUAUUGUAGUUAAAAGUUCCGGCUAGGCCGUGAAAAAUUGACUUAGGGAGUGUGCUUUUCACCA .((..(....)..)).......((((((((..((((((((..((((((((((.(((...(((......)---------------)))))))))))))))(((((...(((((((...)))).)))...))))))))))))).))))))))..... ( -38.50, z-score = -3.09, R) >droSim1.chr2R 5677951 155 + 19596830 CGUGGCGUAAGAAACAUACUUAAAAGGCACUCCCUCUUAAAUUACAAUAAUUUAUUACAGUCUAACUUGUCUUUUACUGCACUGGCAAUAAAUAUUGUAGUUAAAAGUUCCGGCUAGGCCGUGAAAAAUUGACUAAGGGAGUGUGCUUUUAACUA .((..(....)..))....((((((((((((((((.......(((((((.((((((.(((((................).))))..)))))))))))))(((((...(((((((...)))).)))...)))))..))))))))).)))))))... ( -44.29, z-score = -3.65, R) >droSec1.super_1 4699395 141 + 14215200 CGUGGCGUAAGAAACAUACUUAAAAGGCACUCCCUCUUAAAUUACAAUAAUUUAUUACAGUCUAACUUGUCUUUUACUGCACUGGCAAUAAAUAUUGUAGUUAAGAGUUCCGGCUAGGC--------------UAAGGGUGUGUGCUUUUAACUA .((..(....)..))....(((((((((((.((((..(((((((...)))))))....((((((.((.(.(((((((((((.((........)).))))).))))))..).)).)))))--------------).)))).)))).)))))))... ( -35.10, z-score = -1.52, R) >consensus CGUGGCGUAAGAAACAUACUUAAAAGGCACUCCCUCUUAAAUUACAAUAAUUUAUUACAGUCUAACUUGUCUUUUACUGCACUGGCAAUAAAUAUUGUAGUUAAAAGUUCCGGCUAGGCCGUGAAAAAUUGACUAAGGGAGUGUGCUUUUAACUA .((..(....)..))....((((((((((((((((....((((((((((.((((((...(((.....................)))))))))))))))))))........((((...))))..............))))))))).)))))))... (-27.05 = -30.50 + 3.45)
| Location | 7,130,572 – 7,130,712 |
|---|---|
| Length | 140 |
| Sequences | 3 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 84.52 |
| Shannon entropy | 0.21328 |
| G+C content | 0.35789 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -26.41 |
| Energy contribution | -27.64 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7130572 140 - 21146708 UGGUGAAAAGCACACUCCCUAAGUCAAUUUUUCACGGCCUAGCCGGAACUUUUAACUACAAUAUUUAUUGC---------------CAAGUUAGACUGUAAUAAAAUAUUGUAAUUUAAGAAGGAGUGCCUUUUAAGUAUGUUUCUUACGCCACG ((((((((((..((((((............(((.((((...))))))).((((((.(((((((((((((((---------------..((.....))))))).))))))))))..)))))).)))))).)))))(((.......))).))))).. ( -35.00, z-score = -2.84, R) >droSim1.chr2R 5677951 155 - 19596830 UAGUUAAAAGCACACUCCCUUAGUCAAUUUUUCACGGCCUAGCCGGAACUUUUAACUACAAUAUUUAUUGCCAGUGCAGUAAAAGACAAGUUAGACUGUAAUAAAUUAUUGUAAUUUAAGAGGGAGUGCCUUUUAAGUAUGUUUCUUACGCCACG ((.(((((((..(((((((((((((.....(((.((((...)))))))....(((((......((((((((....)))))))).....)))))))))....(((((((...))))))).))))))))).))))))).))................ ( -41.20, z-score = -3.41, R) >droSec1.super_1 4699395 141 - 14215200 UAGUUAAAAGCACACACCCUUA--------------GCCUAGCCGGAACUCUUAACUACAAUAUUUAUUGCCAGUGCAGUAAAAGACAAGUUAGACUGUAAUAAAUUAUUGUAAUUUAAGAGGGAGUGCCUUUUAAGUAUGUUUCUUACGCCACG ((.(((((((..(((.(((...--------------.((.....))...((((((.(((((((((((((((.(((...................))))))))))).)))))))..))))))))).))).))))))).))................ ( -29.51, z-score = -0.82, R) >consensus UAGUUAAAAGCACACUCCCUUAGUCAAUUUUUCACGGCCUAGCCGGAACUUUUAACUACAAUAUUUAUUGCCAGUGCAGUAAAAGACAAGUUAGACUGUAAUAAAUUAUUGUAAUUUAAGAGGGAGUGCCUUUUAAGUAUGUUUCUUACGCCACG ((.(((((((..(((((((...............((((...))))....((((((.(((((((((((((((..........................)))))))).)))))))..))))))))))))).))))))).))................ (-26.41 = -27.64 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:53 2011