
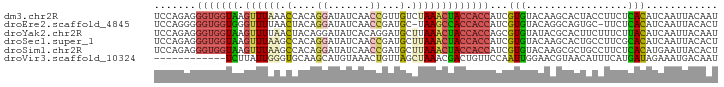
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,118,775 – 7,118,869 |
| Length | 94 |
| Max. P | 0.993658 |

| Location | 7,118,775 – 7,118,869 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.57492 |
| G+C content | 0.42770 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
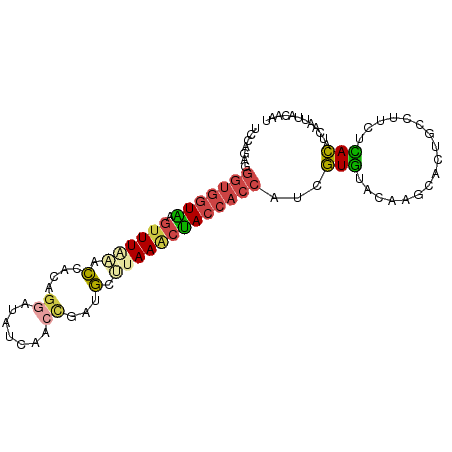
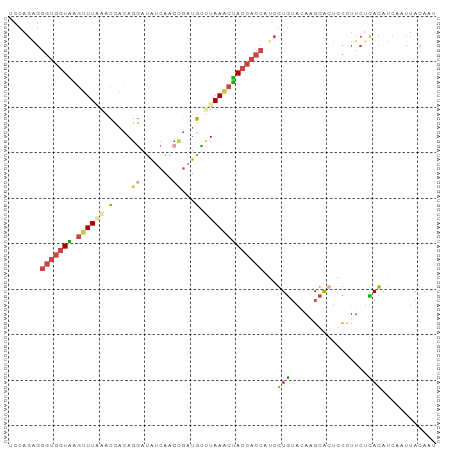
>dm3.chr2R 7118775 94 + 21146708 UCCAGAGGGUGGUAAGUUUAAACCACAGGAUAUCAACCGUUGUCUAAACUACCACCAUCGUGUACAAGCACUACCUUCUCACAUCAAUUACAAU ....((((((((((.(((((.((.((.((.......)))).)).))))))))))))...((((....))))......))).............. ( -24.80, z-score = -3.70, R) >droEre2.scaffold_4845 3912460 92 - 22589142 UCCAGGGGGUGGUGGGUUUUAACUACAGGAUAUCAACCGAUGC-UAAGCCACCACCAUCGUGUACAGGCAGUGC-UUCUCACAUCAAUUACACU ....(..((((((((((.......)).((.((((....)))))-)...))))))))..)(((((...(..(((.-....)))..)...))))). ( -23.50, z-score = -0.28, R) >droYak2.chr2R 19113420 94 - 21139217 UCCAGAGGGUGGUAAGUUUUAACUACAGGAUAUCACAGGAUGCUUAAACUACCACCAGCGUGUAUACGCACUUCUUUCUUACAUCAAUUACAAU ...(((((((((((.((((.......(((.((((....)))))))))))))))))).((((....))))......))))............... ( -19.91, z-score = -1.67, R) >droSec1.super_1 4687516 94 + 14215200 UCCAGAGGGUGGUAAGUUUAAGCCACAGGAUAUCAACCGAUGCUUAAACUACCACCAUCGUGUACAAGCACUGCCUUCGCACAUCAAUUACACU ....((.(((((((.((((((((....((.......))...))))))))))))))).))(((((.......(((....))).......))))). ( -30.34, z-score = -4.57, R) >droSim1.chr2R 5665808 94 + 19596830 UCCAGAGGGUGGUAAGUUUAAGCCACAGGAUAUCAACCGAUGCUUAAACUACCACCAUCGUGUACAAGCGCUGCCUUCUCACAUGAAUUACACU ....((((((((((.((((((((....((.......))...)))))))))))))))...((((....))))......))).............. ( -29.00, z-score = -3.49, R) >droVir3.scaffold_10324 292001 82 + 1288806 ------------UCUUAUUGGGUGCAAGCAUGUAAACUGUUAGCUAAACGACUGUUCCAAUUGGAACGUAACAUUUCAUGAUAGAAAUGACAAU ------------.....(((..(((.((((.(....))))).)).)..)))..(((((....)))))....((((((......))))))..... ( -16.30, z-score = -0.98, R) >consensus UCCAGAGGGUGGUAAGUUUAAACCACAGGAUAUCAACCGAUGCUUAAACUACCACCAUCGUGUACAAGCACUGCCUUCUCACAUCAAUUACAAU .......(((((((.((((((.(....((.......))...).)))))))))))))...(((.................)))............ (-10.98 = -11.40 + 0.42)
| Location | 7,118,775 – 7,118,869 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.57492 |
| G+C content | 0.42770 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -11.67 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
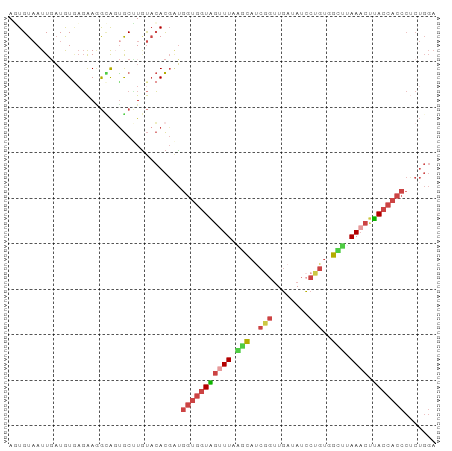
>dm3.chr2R 7118775 94 - 21146708 AUUGUAAUUGAUGUGAGAAGGUAGUGCUUGUACACGAUGGUGGUAGUUUAGACAACGGUUGAUAUCCUGUGGUUUAAACUUACCACCCUCUGGA ............((((.(((......))).).)))((.(((((((((((((((.((((........)))).)))))))).))))))).)).... ( -28.90, z-score = -2.35, R) >droEre2.scaffold_4845 3912460 92 + 22589142 AGUGUAAUUGAUGUGAGAA-GCACUGCCUGUACACGAUGGUGGUGGCUUA-GCAUCGGUUGAUAUCCUGUAGUUAAAACCCACCACCCCCUGGA .(((((......(((....-.)))......)))))...((((((((.(((-((((.((.......)).)).)))))...))))))))....... ( -26.80, z-score = -1.14, R) >droYak2.chr2R 19113420 94 + 21139217 AUUGUAAUUGAUGUAAGAAAGAAGUGCGUAUACACGCUGGUGGUAGUUUAAGCAUCCUGUGAUAUCCUGUAGUUAAAACUUACCACCCUCUGGA ...................(((...((((....)))).(((((((((((.(((..................))).)))).))))))).)))... ( -17.97, z-score = 0.16, R) >droSec1.super_1 4687516 94 - 14215200 AGUGUAAUUGAUGUGCGAAGGCAGUGCUUGUACACGAUGGUGGUAGUUUAAGCAUCGGUUGAUAUCCUGUGGCUUAAACUUACCACCCUCUGGA .(((((...(...(((....)))...)...)))))((.(((((((((((((((..(((........)))..)))))))).))))))).)).... ( -36.40, z-score = -3.88, R) >droSim1.chr2R 5665808 94 - 19596830 AGUGUAAUUCAUGUGAGAAGGCAGCGCUUGUACACGAUGGUGGUAGUUUAAGCAUCGGUUGAUAUCCUGUGGCUUAAACUUACCACCCUCUGGA .(((((.....(((......))).......)))))((.(((((((((((((((..(((........)))..)))))))).))))))).)).... ( -31.50, z-score = -2.25, R) >droVir3.scaffold_10324 292001 82 - 1288806 AUUGUCAUUUCUAUCAUGAAAUGUUACGUUCCAAUUGGAACAGUCGUUUAGCUAACAGUUUACAUGCUUGCACCCAAUAAGA------------ ((((.((((((......))))))....(((((....)))))....((..(((.............))).))...))))....------------ ( -14.42, z-score = -0.87, R) >consensus AGUGUAAUUGAUGUGAGAAGGCAGUGCUUGUACACGAUGGUGGUAGUUUAAGCAUCGGUUGAUAUCCUGUGGCUUAAACUUACCACCCUCUGGA ......................................(((((((((((.(((..(((........)))..))).)))).)))))))....... (-11.67 = -12.40 + 0.73)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:50 2011