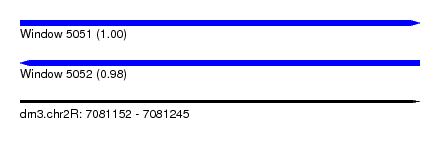
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,081,152 – 7,081,245 |
| Length | 93 |
| Max. P | 0.996780 |

| Location | 7,081,152 – 7,081,245 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.40588 |
| G+C content | 0.51211 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -28.12 |
| Energy contribution | -29.30 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
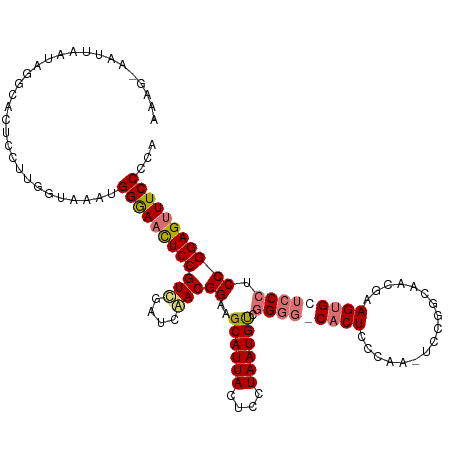
>dm3.chr2R 7081152 93 + 21146708 ---------------------------ACUGGAAACUCCGUCGAUCGACGGAAGCAUUACUCCUAAUGUCGGGGGCACUCCCAAAUCCAUCAGAGAAGUGCUCCCUCCGGAGUUUCCCCA ---------------------------...((((((((((((....)))(((.((((((....)))))).(((((((((.((..........).).)))))))))))))))))))))... ( -39.50, z-score = -4.43, R) >droEre2.scaffold_4845 3874804 118 - 22589142 AUAAUUAUUAAUAGGCACUCCUUGAUAAAUGGGAACUCCGUCGAUCGACGGAAGCAUUACUCGUAAUGUUGGGG-CACUCCCAA-UCCGGCAACGAAGUGCUCCACCCGGAGCUUCCCCA ....((((((..(((....)))))))))..((((((((((((....)))((.(((((((....)))))))((((-((((.....-..((....)).))))))))..)))))).))))).. ( -41.10, z-score = -3.16, R) >droYak2.chr2R 19076631 117 - 21139217 AAAGAAAUUAAUAGGCACUCGUUGGCAAAUGGGAAUUCCGUCGAUCAACGGAAGCAUUACUCGUAAUGUCGGA--CACUCCCAA-UCCGGCCACGAAGUGCUCCCACCGGAGUUUCCCCA ...((((((....(((((((((.(((...(((((.((((((......))))))((((((....))))))....--...))))).-....)))))).))))))((....)))))))).... ( -37.30, z-score = -2.28, R) >droSim1.chr2R 5626865 119 + 19596830 AAUGGAAUUAAUAGGCACUCCUUGGUAAAUGGGAACUCCGUUGAUCAACGGAAGCAUUACUCCUAAUGCCGGGGGCACUCCCAA-UUCUUCAACAAAGGGCUCCCUCCGGAGUUUCCCCA ..(((..(((..(((....)))...)))..((((((((((((((.....((((((((((....)))))).((((....))))..-)))))))))..(((....)))..)))))))))))) ( -37.00, z-score = -0.93, R) >consensus AAAG_AAUUAAUAGGCACUCCUUGGUAAAUGGGAACUCCGUCGAUCAACGGAAGCAUUACUCCUAAUGUCGGGG_CACUCCCAA_UCCGGCAACGAAGUGCUCCCUCCGGAGUUUCCCCA ..............................((((((((((((....)))((..((((((....)))))).(((((((((.................))))))))).)))))))))))... (-28.12 = -29.30 + 1.19)

| Location | 7,081,152 – 7,081,245 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.40588 |
| G+C content | 0.51211 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -30.26 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
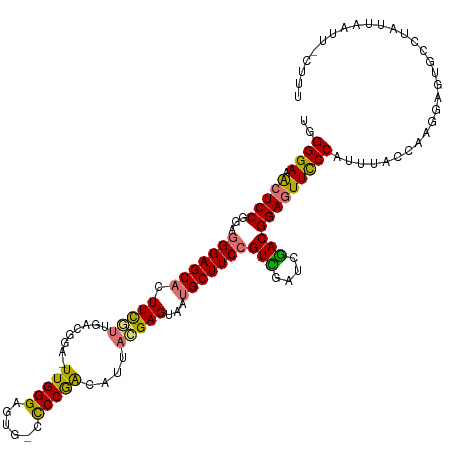
>dm3.chr2R 7081152 93 - 21146708 UGGGGAAACUCCGGAGGGAGCACUUCUCUGAUGGAUUUGGGAGUGCCCCCGACAUUAGGAGUAAUGCUUCCGUCGAUCGACGGAGUUUCCAGU--------------------------- ...(((((((((...(((((((...((((((((...(((((......)))))))))).)))...)))))))(((....))))))))))))...--------------------------- ( -43.20, z-score = -3.62, R) >droEre2.scaffold_4845 3874804 118 + 22589142 UGGGGAAGCUCCGGGUGGAGCACUUCGUUGCCGGA-UUGGGAGUG-CCCCAACAUUACGAGUAAUGCUUCCGUCGAUCGACGGAGUUCCCAUUUAUCAAGGAGUGCCUAUUAAUAAUUAU .((((...)))).(((((.(((((((.(((...((-.((((((..-......(((((....))))).(((((((....))))))))))))).))..)))))))))))))))......... ( -39.60, z-score = -1.37, R) >droYak2.chr2R 19076631 117 + 21139217 UGGGGAAACUCCGGUGGGAGCACUUCGUGGCCGGA-UUGGGAGUG--UCCGACAUUACGAGUAAUGCUUCCGUUGAUCGACGGAAUUCCCAUUUGCCAACGAGUGCCUAUUAAUUUCUUU .(((....).))(((((..(((((.((((((..((-.(((((((.--...(.(((((....)))))).((((((....))))))))))))).))))).)))))))))))))......... ( -40.90, z-score = -1.71, R) >droSim1.chr2R 5626865 119 - 19596830 UGGGGAAACUCCGGAGGGAGCCCUUUGUUGAAGAA-UUGGGAGUGCCCCCGGCAUUAGGAGUAAUGCUUCCGUUGAUCAACGGAGUUCCCAUUUACCAAGGAGUGCCUAUUAAUUCCAUU ..(((((.(((((((((....)))))(((((....-..(((......)))(((((((....)))))))........)))))))))))))).........(((((........)))))... ( -40.70, z-score = -1.30, R) >consensus UGGGGAAACUCCGGAGGGAGCACUUCGUUGACGGA_UUGGGAGUG_CCCCGACAUUACGAGUAAUGCUUCCGUCGAUCGACGGAGUUCCCAUUUACCAAGGAGUGCCUAUUAAUU_CUUU ..((((.(((((...(((((((.(((((........(((((......)))))....)))))...)))))))(((....)))))))))))).............................. (-30.26 = -29.70 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:47 2011