
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,071,167 – 7,071,261 |
| Length | 94 |
| Max. P | 0.597009 |

| Location | 7,071,167 – 7,071,261 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Shannon entropy | 0.52533 |
| G+C content | 0.60832 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -17.77 |
| Energy contribution | -17.69 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.597009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 7071167 94 + 21146708 CCACCGUCCAACUGGCAA-CGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG-CGACGGCGGCGUUUUCAAAGCGA--- ....(((.....(((.((-(((((((.((.(((...(((((.....((((((...))))))..))))).)))-.)).))))))))).)))..))).--- ( -34.70, z-score = -1.32, R) >droSim1.chr2R 5615641 94 + 19596830 CCCCCGUCCAACUGGCAG-CGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG-CGACGGCGGCGUUUUCAAAGCGA--- ....(((.....(((.((-(((((((.((.(((...(((((.....((((((...))))))..))))).)))-.)).))))))))).)))..))).--- ( -34.70, z-score = -0.93, R) >droSec1.super_1 4640875 94 + 14215200 CCCCCGUCCAACUGGCAG-CGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG-CGACGGCGGCGUUUUCAAAGCGA--- ....(((.....(((.((-(((((((.((.(((...(((((.....((((((...))))))..))))).)))-.)).))))))))).)))..))).--- ( -34.70, z-score = -0.93, R) >droYak2.chr2R 19066463 92 - 21139217 --CCCCUCCAACUGGCAA-CGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG-CGACGGCGGCGAUUUCAAAGCGA--- --.(((.((....(....-)(((((((....((((.(((((((((....))))..)))))...))))..)))-)))))).)).)............--- ( -30.50, z-score = -0.64, R) >droEre2.scaffold_4845 3864492 95 - 22589142 CCCCUCUCCAACUGGCAAACGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG-CGACGGCGGCGUUUUCAAAGCGA--- ............(((.((((((((((.((.(((...(((((.....((((((...))))))..))))).)))-.)).)))))))))))))......--- ( -34.00, z-score = -1.84, R) >droAna3.scaffold_13266 786477 97 + 19884421 ACCACCACCGGCCACCAC-UGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGGUGUCUGG-CGACGGCGGCGUUUUCAAAGGCCCAA .........((((.((.(-((((((((.(.((((..(((((((((....))))..))))).)))).)..)))-)))))).)).(....)...))))... ( -38.60, z-score = -1.96, R) >dp4.chr3 1082571 95 - 19779522 CCGACACCCCCCCGCCAC-CACUGCCACGACAGUCGUCUGGCGACGGGCGACGUUGUCGUCUUGCUGUCUGGUCGAUGGCGGUACUUUCAAAGUGA--- ...........((((((.-(...((((.((((((.(((((....)))))((((....))))..)))))))))).).))))))((((.....)))).--- ( -36.30, z-score = -0.97, R) >droPer1.super_34 273323 95 - 916997 CCGACACCCCCCCGCCAC-CACUGCCACGACAGUCGUCUGGCGACGGGCGACGUUGUCGUCUUGCUGUCUGGUCGAUGGCGGUAUUUUCAAAGCGA--- ...........((((((.-(...((((.((((((.(((((....)))))((((....))))..)))))))))).).))))))..............--- ( -35.80, z-score = -0.90, R) >droVir3.scaffold_12823 1200063 92 - 2474545 ---CUACCCCGUAGCUUAGCUCCGCCUCGGACUUGGCCAACUGUCGAUGCAAGUCGUCGUCUCGAUGUCUGC-AGUCGGCGGCCUUUUCAAAGUGA--- ---.....(((..((........))..)))....((((....(((((((((.(.(((((...))))).))))-).)))))))))............--- ( -28.70, z-score = -0.87, R) >consensus CCCCCAUCCAACUGGCAA_CGUCGCCAUCUCCAACUGCGACUGUCUGGCGACGAUGUCGUCUUGUUGUCUGG_CGACGGCGGCGUUUUCAAAGCGA___ ....................((((((....................((((((...)))))).....(((.....)))))))))................ (-17.77 = -17.69 + -0.08)

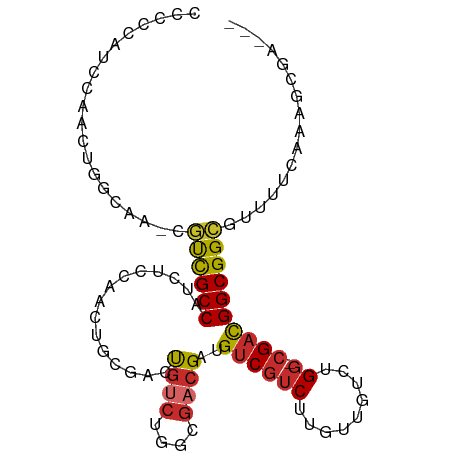
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:45 2011