| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,066,893 – 7,066,954 |
| Length | 61 |
| Max. P | 0.998530 |

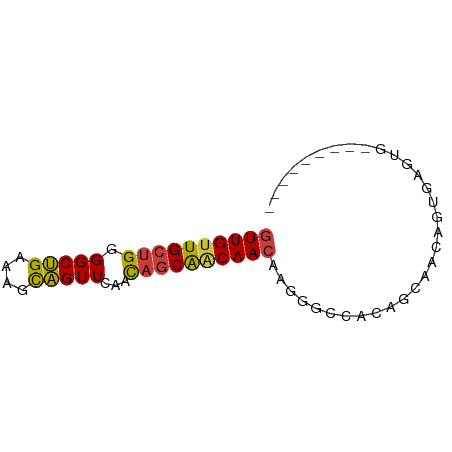
| Location | 7,066,893 – 7,066,954 |
|---|---|
| Length | 61 |
| Sequences | 7 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 72.06 |
| Shannon entropy | 0.53012 |
| G+C content | 0.51217 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -16.32 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998530 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 7066893 61 + 21146708 GUUGUUGCUGUGGCUGAAAGCAGUUCACUAGCAACAACAAGGGCCACAGCAACAGUGAGUG--------- .((((((((((((((....((((....)).))..(.....)))))))))))))))......--------- ( -24.60, z-score = -2.46, R) >droSim1.chr2R 5611433 61 + 19596830 GUUGUUGCUGGGGCUGAGAGCAGUUUCCUAGCAACAACAAGGACCACAGCAACAGUGAGUG--------- ((((((((((((((((....))))..))))))))))))....(((((.......))).)).--------- ( -25.30, z-score = -3.05, R) >droSec1.super_1 4636475 61 + 14215200 GUUGUUGCUGUGGCUGAAAGCAGUUCCUUAGCAACAACAAGGACCACAGCAACAGUGAGUG--------- .(((((((((((((((....))).(((((.(......))))))))))))))))))......--------- ( -23.30, z-score = -2.49, R) >droYak2.chr2R 19062074 61 - 21139217 GUUGUUGCUGUGGCUGAAAGCAGUUCAGCAGCAACAACAAGGGCCACAGCAACAGUGAGUG--------- (((((((((((.((((....))))...)))))))))))....(((((.......))).)).--------- ( -24.10, z-score = -2.11, R) >droEre2.scaffold_4845 3860349 61 - 22589142 GUUGUUGCUGCGGCUGAAAGCAGUUCAGCAGCAACAACAAGGGCCACAGCAACAGUAAGUG--------- (((((((((((.((((....))))...)))))))))))....((....))...........--------- ( -23.70, z-score = -2.58, R) >droPer1.super_34 268953 70 - 916997 GUUGUUGCCGGGGCAGAAAACUGUUCAACAGCGCCAACAAAGCAGGCAGAGGCAGAGGCAACGACGAGUG (((((((((.((((((....))))))......(((.........))).........)))))))))..... ( -25.50, z-score = -2.57, R) >droWil1.scaffold_180697 3439711 50 + 4168966 -UUGUUGCUGGGGCAGAAAGUUGUUCAACAACAGCAACAACAU-GACGUUCA------------------ -(((((((((..(..(((.....)))..)..)))))))))...-........------------------ ( -14.30, z-score = -1.67, R) >consensus GUUGUUGCUGGGGCUGAAAGCAGUUCAACAGCAACAACAAGGGCCACAGCAACAGUGAGUG_________ ((((((((((.(((((....)))))...))))))))))................................ (-16.32 = -15.90 + -0.42)

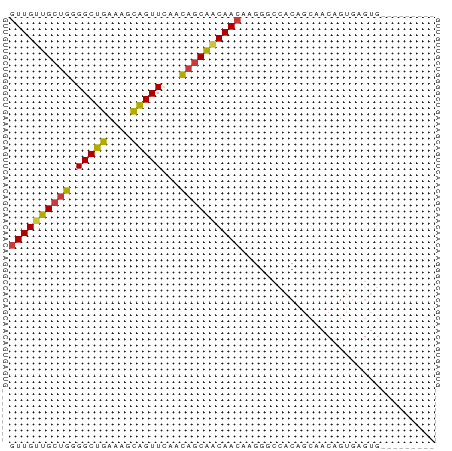
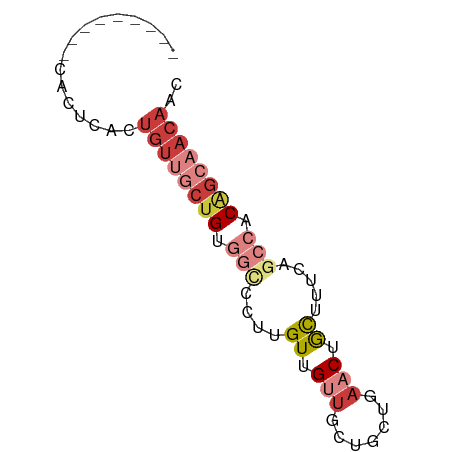
| Location | 7,066,893 – 7,066,954 |
|---|---|
| Length | 61 |
| Sequences | 7 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 72.06 |
| Shannon entropy | 0.53012 |
| G+C content | 0.51217 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -8.69 |
| Energy contribution | -10.80 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980965 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 7066893 61 - 21146708 ---------CACUCACUGUUGCUGUGGCCCUUGUUGUUGCUAGUGAACUGCUUUCAGCCACAGCAACAAC ---------.......((((((((((((...((.....((.((....))))...)))))))))))))).. ( -24.40, z-score = -3.15, R) >droSim1.chr2R 5611433 61 - 19596830 ---------CACUCACUGUUGCUGUGGUCCUUGUUGUUGCUAGGAAACUGCUCUCAGCCCCAGCAACAAC ---------.......((((((((.((.....((((..((.((....))))...)))))))))))))).. ( -22.00, z-score = -2.80, R) >droSec1.super_1 4636475 61 - 14215200 ---------CACUCACUGUUGCUGUGGUCCUUGUUGUUGCUAAGGAACUGCUUUCAGCCACAGCAACAAC ---------.......((((((((((((((((((....)).))))).(((....)))))))))))))).. ( -24.30, z-score = -3.31, R) >droYak2.chr2R 19062074 61 + 21139217 ---------CACUCACUGUUGCUGUGGCCCUUGUUGUUGCUGCUGAACUGCUUUCAGCCACAGCAACAAC ---------.......((((((((((((....((.(((.(....)))).)).....)))))))))))).. ( -23.60, z-score = -2.93, R) >droEre2.scaffold_4845 3860349 61 + 22589142 ---------CACUUACUGUUGCUGUGGCCCUUGUUGUUGCUGCUGAACUGCUUUCAGCCGCAGCAACAAC ---------.......((((((((((((....((.(((.(....)))).)).....)))))))))))).. ( -23.20, z-score = -2.98, R) >droPer1.super_34 268953 70 + 916997 CACUCGUCGUUGCCUCUGCCUCUGCCUGCUUUGUUGGCGCUGUUGAACAGUUUUCUGCCCCGGCAACAAC ........((((((...((....(((.((...)).)))((((.....)))).....))...))))))... ( -18.50, z-score = -1.12, R) >droWil1.scaffold_180697 3439711 50 - 4168966 ------------------UGAACGUC-AUGUUGUUGCUGUUGUUGAACAACUUUCUGCCCCAGCAACAA- ------------------........-...(((((((((..((.(((.....))).))..)))))))))- ( -13.10, z-score = -1.90, R) >consensus _________CACUCACUGUUGCUGUGGCCCUUGUUGUUGCUGCUGAACUGCUUUCAGCCACAGCAACAAC ................((((((((.(((....((.(((.......))).)).....))).)))))))).. ( -8.69 = -10.80 + 2.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:44 2011