| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,059,732 – 7,059,843 |
| Length | 111 |
| Max. P | 0.998920 |

| Location | 7,059,732 – 7,059,843 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.48 |
| Shannon entropy | 0.58759 |
| G+C content | 0.53156 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -15.87 |
| Energy contribution | -18.07 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
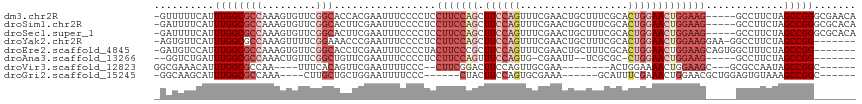
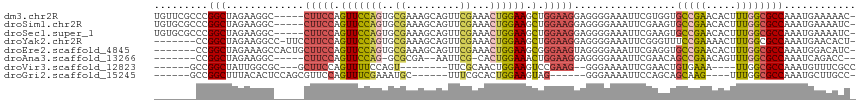
>dm3.chr2R 7059732 111 + 21146708 -GUUUUUCAUUUGGCGCCAAAGUGUUCGGCACCACGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAG-----GCCUUCUAGCCGGGCGAACA -((((....((((....))))..(((((((.....(.....)....((((((((.(((((((..((((.....)))).))))))))))))))-----).......))))))))))). ( -38.80, z-score = -2.78, R) >droSim1.chr2R 5602519 111 + 19596830 -GAUUUUCAUUUGGCGCCAAAGUGUUCGGCACUUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAG-----GCCUUCUAGCCGGGCGCACA -........((((....))))(((((((((.....(.....)....((((((((.(((((((..((((.....)))).))))))))))))))-----).......)))))))))... ( -41.50, z-score = -3.56, R) >droSec1.super_1 4629246 111 + 14215200 -GAUUUUCAUUUGGCGCCAAAGUGUUCGGCACUUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAG-----GCCUUCUAGCCGGGCGCACA -........((((....))))(((((((((.....(.....)....((((((((.(((((((..((((.....)))).))))))))))))))-----).......)))))))))... ( -41.50, z-score = -3.56, R) >droYak2.chr2R 19054609 108 - 21139217 -AGUGUUCAUUUGGCGCCAAAGUUUUCGGAAACCCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAGGAA-GGCCUUCUAGCCGG------- -..........((((.........(((((....)))))....((.(((((((((.(((((((..((((.....)))).)))))))))))))))).-)).......)))).------- ( -41.30, z-score = -3.59, R) >droEre2.scaffold_4845 3848927 109 - 22589142 -GAUGUCCAUUUGGCGCCAAAGUGUUCGGCACCUCGAAUUUCCCCUACUUCCCGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAGCAGUGGCUUUCUAGCCGG------- -...........((.(((.........))).))..................(((((((((((((((...(((....))).)))))))))))))...((((....))))))------- ( -36.20, z-score = -2.38, R) >droAna3.scaffold_13266 776282 99 + 19884421 --GGUCUGAUUUGGCGCCAAACUGUUCGGCUGUUCGAAUUUCCCCUCCUUCCAGUUUCCAGUG-CGAAUU--UCGCGC-CUGGAACUGGAAG-----GCCUUCUAGCCGG------- --(((..((...((((((.........))).................(((((((((.((((((-((....--.)))).-)))))))))))))-----))).))..)))..------- ( -40.40, z-score = -3.87, R) >droVir3.scaffold_12823 1183543 94 - 2474545 GGCGAAACAUUUGGCGCCAA----UUUCACAGUUCGAAUUUUCCC--CUUCGGACUUCCAGUUGCGAA--------ACUGGAAAACUGGAAGC---GCGCCAAUAGCCGGC------ (((.......(((((((...----(((((..(((((((.......--.)))))))((((((((....)--------)))))))...)))))..---)))))))..)))...------ ( -36.40, z-score = -3.53, R) >droGri2.scaffold_15245 5937965 94 + 18325388 -GGCAAGCAUUUGGCGCCAAA----CUUGCUGCUGGAAUUUUCCC------CUACUUCCAGUGCGAAA------GCAUUUCGAAACUGGAACGCUGGAGUGUAAAGCCGGC------ -((((((..((((....))))----))))))(((((....((((.------....(((((((.((((.------....))))..)))))))....)))).......)))))------ ( -29.50, z-score = -0.72, R) >consensus _GAUUUUCAUUUGGCGCCAAAGUGUUCGGCACCUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCACUGGAACUGGAAG_____GCCUUCUAGCCGG_______ ..........((((((((.........))).................(((((((.((((((..................))))))))))))).............)))))....... (-15.87 = -18.07 + 2.20)
| Location | 7,059,732 – 7,059,843 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.48 |
| Shannon entropy | 0.58759 |
| G+C content | 0.53156 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.89 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.998920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
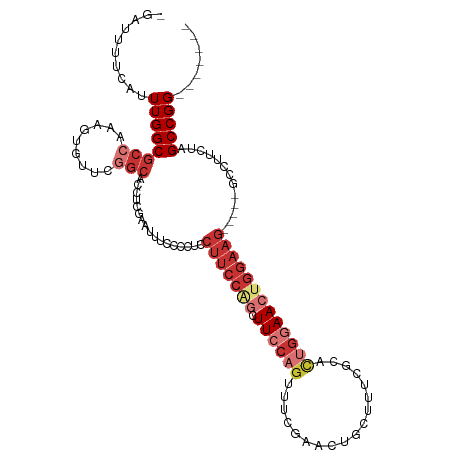
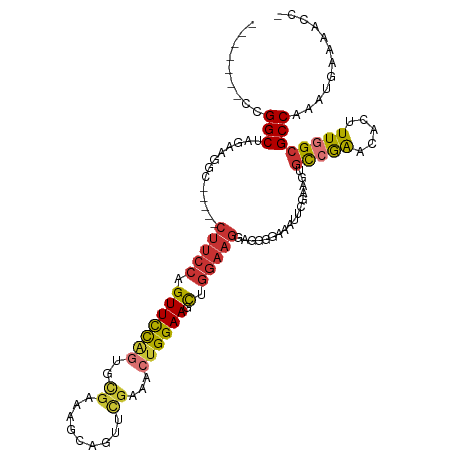
>dm3.chr2R 7059732 111 - 21146708 UGUUCGCCCGGCUAGAAGGC-----CUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGUGGUGCCGAACACUUUGGCGCCAAAUGAAAAAC- ..(((.(((((((....)))-----)(((((((((((((.((((.....))))..))))).))))))))...)))))).(((.(((((((((.....)))))))))...)))....- ( -47.90, z-score = -4.06, R) >droSim1.chr2R 5602519 111 - 19596830 UGUGCGCCCGGCUAGAAGGC-----CUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCCAAAUGAAAAUC- .((((.(((((((....)))-----)(((((((((((((.((((.....))))..))))).))))))))...)))......(((((((.....)))))))))))............- ( -43.20, z-score = -2.91, R) >droSec1.super_1 4629246 111 - 14215200 UGUGCGCCCGGCUAGAAGGC-----CUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCCAAAUGAAAAUC- .((((.(((((((....)))-----)(((((((((((((.((((.....))))..))))).))))))))...)))......(((((((.....)))))))))))............- ( -43.20, z-score = -2.91, R) >droYak2.chr2R 19054609 108 + 21139217 -------CCGGCUAGAAGGCC-UUCCUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGGGUUUCCGAAAACUUUGGCGCCAAAUGAACACU- -------..(((..((((.((-(((((((((((((((((.((((.....))))..))))).))))))))))))))....(((((....)))))..))))...)))...........- ( -48.90, z-score = -4.45, R) >droEre2.scaffold_4845 3848927 109 + 22589142 -------CCGGCUAGAAAGCCACUGCUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCGGGAAGUAGGGGAAAUUCGAGGUGCCGAACACUUUGGCGCCAAAUGGACAUC- -------((((((....)))).(((((((((((((.(.(((....))).)..).))))))))))))......))..........((((((((.....))))))))...........- ( -46.90, z-score = -4.28, R) >droAna3.scaffold_13266 776282 99 - 19884421 -------CCGGCUAGAAGGC-----CUUCCAGUUCCAG-GCGCGA--AAUUCG-CACUGGAAACUGGAAGGAGGGGAAAUUCGAACAGCCGAACAGUUUGGCGCCAAAUCAGACC-- -------..(((.......(-----(((((((((((((-..(((.--....))-).)))).))))))))))................(((((.....))))))))..........-- ( -40.00, z-score = -3.34, R) >droVir3.scaffold_12823 1183543 94 + 2474545 ------GCCGGCUAUUGGCGC---GCUUCCAGUUUUCCAGU--------UUCGCAACUGGAAGUCCGAAG--GGGAAAAUUCGAACUGUGAAA----UUGGCGCCAAAUGUUUCGCC ------((.(((..(((((((---..(((((((((((((((--------(....)))))))..(((....--))).......)))))).))).----...)))))))..)))..)). ( -32.60, z-score = -1.54, R) >droGri2.scaffold_15245 5937965 94 - 18325388 ------GCCGGCUUUACACUCCAGCGUUCCAGUUUCGAAAUGC------UUUCGCACUGGAAGUAG------GGGAAAAUUCCAGCAGCAAG----UUUGGCGCCAAAUGCUUGCC- ------((.((..((...((((.((.(((((((..((((....------.)))).))))))))).)------)))..))..)).)).(((((----((((....)))..)))))).- ( -30.60, z-score = -1.17, R) >consensus _______CCGGCUAGAAGGC_____CUUCCAGUUCCAGUGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCCAAAUGAAAACC_ .........(((.............(((((.(((((((..((.........))...)))))).).))))).................(((((.....))))))))............ (-18.43 = -18.89 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:42 2011