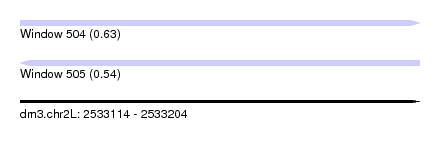
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,533,114 – 2,533,204 |
| Length | 90 |
| Max. P | 0.634309 |

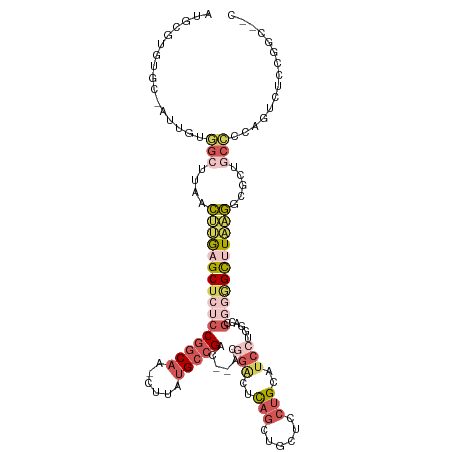
| Location | 2,533,114 – 2,533,204 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.62918 |
| G+C content | 0.58745 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -15.56 |
| Energy contribution | -18.32 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
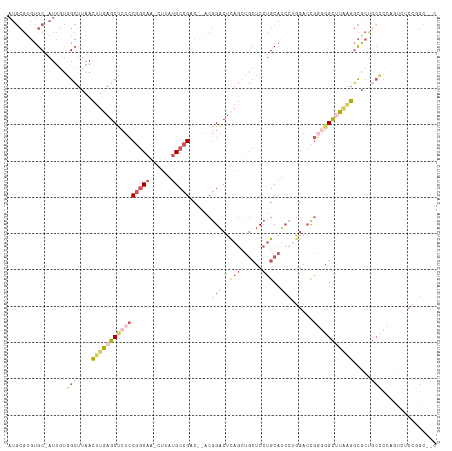
>dm3.chr2L 2533114 90 + 23011544 AUGCGUGUGC-AUUGUAGCUUAACUUGAGCUCUCCGGCAA-CUUAUGCCGAC--ACGGACUCAGCUGCUCCUGCA------GGUGCCAGUCUCCGGCACC------------------ .......(((-(..((((((......(((.(((.(((((.-....)))))..--..))))))))))))...))))------((((((.......))))))------------------ ( -31.40, z-score = -1.15, R) >droYak2.chr2L 2525464 114 + 22324452 AUGCGUGUGC-AUUCUGGCUUAACUUGAGCUCUCCGGCAA-CUUAUGCCGAC--ACGGACUCAGAUGCUCCUGCAUCCUGGACUGGGGGCUUAAGGCACUGCCCCAGUGCCCGGCUCC ..((.((.((-(((..(((....((((((((((.(((((.-....)))))..--.(((.((..(((((....)))))..)).))))))))))))).....)))..))))).))))... ( -44.40, z-score = -1.82, R) >droSec1.super_5 706979 114 + 5866729 AUGCGUGUGC-AUUGUGGCUUAACUUGAGCUCUCCGGCAA-CUUAUGCCGAC--ACGGACUCAGCUGCUCCUGCAUCCUGGACCGGGGGCUUAAGGCGCUGCCCCAGUCUCCGGCACC ......((((-((((.(((....((((((((((((((((.-....)))))..--..((.(.(((.(((....)))..)))).))))))))))))).....))).)))).....)))). ( -42.40, z-score = -0.65, R) >droSim1.chr2L 2495388 114 + 22036055 AUGCGUGUGC-AUUGUGGCUUAACUUAAGCUCUCCGGCAA-CUUAUGCCGAC--ACGGACUCAGCUGCUCCUGCAUCCUGGACCGGGGGCUUAAGGCGCUGCCCCAGUCUCCGGCAGC ..((....((-......))....((((((((((((((((.-....)))))..--..((.(.(((.(((....)))..)))).)))))))))))))))((((((.........)))))) ( -42.80, z-score = -0.56, R) >droVir3.scaffold_12963 10222759 103 - 20206255 ------CUGCUAUGCCAGGUCAAACAAAGCCACGCAACAAACAAAAGCCGCCCCACAGGCUUAAGACGGCAGGCAUC--AGGCCACAUGCAUUGUCUGAUGCUUCGCAGUC------- ------((((..((((.(((........))).............(((((........))))).....))))((((((--((((..........))))))))))..))))..------- ( -34.50, z-score = -2.26, R) >consensus AUGCGUGUGC_AUUGUGGCUUAACUUGAGCUCUCCGGCAA_CUUAUGCCGAC__ACGGACUCAGCUGCUCCUGCAUCCUGGACCGGGGGCUUAAGGCGCUGCCCCAGUCUCCGGC__C ................(((....((((((((((((((((......)))))......(((..(((......)))..)))......))))))))))).....)))............... (-15.56 = -18.32 + 2.76)

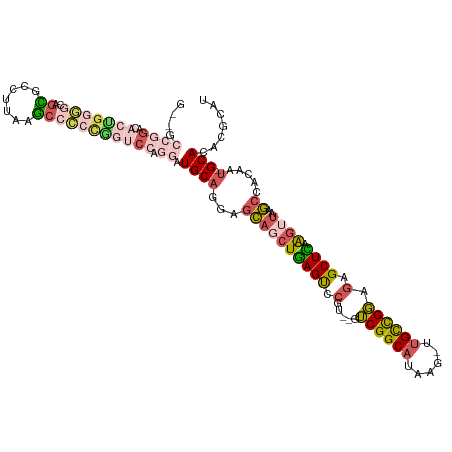
| Location | 2,533,114 – 2,533,204 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 64.17 |
| Shannon entropy | 0.62918 |
| G+C content | 0.58745 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -15.94 |
| Energy contribution | -17.26 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2533114 90 - 23011544 ------------------GGUGCCGGAGACUGGCACC------UGCAGGAGCAGCUGAGUCCGU--GUCGGCAUAAG-UUGCCGGAGAGCUCAAGUUAAGCUACAAU-GCACACGCAU ------------------(((((((....).))))))------((((..((((((((((..(..--.((((((....-.)))))).)..))).))))..)))....)-)))....... ( -38.60, z-score = -2.72, R) >droYak2.chr2L 2525464 114 - 22324452 GGAGCCGGGCACUGGGGCAGUGCCUUAAGCCCCCAGUCCAGGAUGCAGGAGCAUCUGAGUCCGU--GUCGGCAUAAG-UUGCCGGAGAGCUCAAGUUAAGCCAGAAU-GCACACGCAU .((((((((((((((((....((.....)))))))))...((((((....))))))..))))).--.((((((....-.))))))...)))).............((-((....)))) ( -43.80, z-score = -1.26, R) >droSec1.super_5 706979 114 - 5866729 GGUGCCGGAGACUGGGGCAGCGCCUUAAGCCCCCGGUCCAGGAUGCAGGAGCAGCUGAGUCCGU--GUCGGCAUAAG-UUGCCGGAGAGCUCAAGUUAAGCCACAAU-GCACACGCAU .((((..(.((((((((..((.......))))))))))).(..((((.(.(((((((((..(..--.((((((....-.)))))).)..))).))))..)))....)-)))..))))) ( -43.80, z-score = -0.76, R) >droSim1.chr2L 2495388 114 - 22036055 GCUGCCGGAGACUGGGGCAGCGCCUUAAGCCCCCGGUCCAGGAUGCAGGAGCAGCUGAGUCCGU--GUCGGCAUAAG-UUGCCGGAGAGCUUAAGUUAAGCCACAAU-GCACACGCAU (((((((....)...))))))(((((((((((((((.((((..(((....))).))).).))).--...((((....-.)))))).).)))))))....((......-))....)).. ( -42.70, z-score = -0.49, R) >droVir3.scaffold_12963 10222759 103 + 20206255 -------GACUGCGAAGCAUCAGACAAUGCAUGUGGCCU--GAUGCCUGCCGUCUUAAGCCUGUGGGGCGGCUUUUGUUUGUUGCGUGGCUUUGUUUGACCUGGCAUAGCAG------ -------..((((...((.((((((((.(((((..((..--((((...(((((((((......)))))))))...)))).))..))).)).))))))))....))...))))------ ( -39.00, z-score = -1.28, R) >consensus G__GCCGGACACUGGGGCAGCGCCUUAAGCCCCCGGUCCAGGAUGCAGGAGCAGCUGAGUCCGU__GUCGGCAUAAG_UUGCCGGAGAGCUCAAGUUAAGCCACAAU_GCACACGCAU ..........(((((((..((.......)))))))))..........((...((((((((.(.....((((((......)))))).).)))).))))...))................ (-15.94 = -17.26 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:39 2011