| Sequence ID | dm3.chr2R |
|---|---|
| Location | 7,027,701 – 7,027,821 |
| Length | 120 |
| Max. P | 0.564328 |

| Location | 7,027,701 – 7,027,821 |
|---|---|
| Length | 120 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.57026 |
| G+C content | 0.54167 |
| Mean single sequence MFE | -37.94 |
| Consensus MFE | -20.93 |
| Energy contribution | -21.41 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
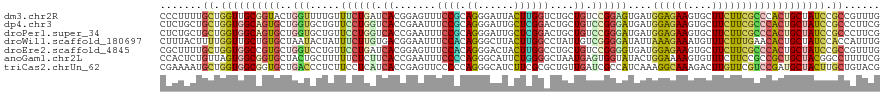
>dm3.chr2R 7027701 120 - 21146708 CCCUUUUGCUGGUUGCGGUACUGGUUUUGUUUCUGAUCACGGAGUUUCCGCAGGGAUUACUUGGUCUGCUGUCCGGAGUGAUGGAGAAGUGCUUCUUCGCCCACUGCUAUCCGCCGUUUG .......((.(((.(((((..(((((........)))))(((.....)))..((((((((((((.(....).)).)))))))((((((....)))))).)))))))).))).))...... ( -33.20, z-score = 0.61, R) >dp4.chr3 1040309 120 + 19779522 CUCUGCUGCUGGUGGCAGUGCUGGUGCUGUUCCUGGUCACCGAAUUUCCGCAGGGAUUGCUCGGACUGCUGUCCGGGAUGAUGGAGAAGUGCUUCUUCGCCCACUGCUAUCCGCCCUUCG .......((.(((((((((..(((((((......)).)))))..........((((((((((((((....))))))).))))((((((....)))))).)))))))))))).))...... ( -44.30, z-score = -0.83, R) >droPer1.super_34 232709 120 + 916997 CUCUGCUGCUGGUGGCAGUGCUGGUGCUGUUCCUGGUCACCGAAUUUCCGCAGGGAUUGCUCGGACUGCUGUCCGGGAUGAUGGAGAAGUGCUUCUUCGCCCACUGCUAUCCGCCCUUCG .......((.(((((((((..(((((((......)).)))))..........((((((((((((((....))))))).))))((((((....)))))).)))))))))))).))...... ( -44.30, z-score = -0.83, R) >droWil1.scaffold_180697 3364494 120 - 4168966 CUUUACUUUUGGUUGCUGUGCUAAUACUAUUUCUUGUGACGGAAUUUCCACAGGGCUUACUUGGCCUAUUGUCGGGGAUAUUAAAGAAAUGUUUCUUUGAACACUGCUAUCCACCAUUUG .........(((..((.(((.......(((..((...(((((.....))...(((((.....)))))...)))))..)))((((((((....)))))))).))).))...)))....... ( -27.70, z-score = -1.66, R) >droEre2.scaffold_4845 3819002 120 + 22589142 CGCUUUUGCUGGUGGCCGUGCUGGUCCUGUUCCUGAUCACGGAGUUUCCACAGGGACUACUUGGCCUGCUGUCCGGGGUGAUGGAGAAGUGCUUCUUCGCCCACUGCUAUCCGCCGUUUG .......((.((((((.((((.((((..(((((((.....((.....)).))))))).....)))).))......((((((.((((.....)))).)))))))).)))))).))...... ( -41.80, z-score = -0.21, R) >anoGam1.chr2L 3641943 120 - 48795086 CCACUCUGUUAGUGGCGGUGCUACUGCUUUUUCUCUUCACCGAAUUUCCCCAGGGCAUUCUGGGGCUAAUGAGUGGUAUACUGGAAAAGUGUUUCUUCCGCCGCUGCUACGGCCUUUUCG .....(((((((((((((....((.((((((((.....((((.(((.(((((((....)))))))..)))...)))).....)))))))))).....)))))))))..))))........ ( -41.90, z-score = -2.80, R) >triCas2.chrUn_62 199158 120 - 341982 CGAAAAUGCUGGUGGCGGUGCUGACCCUCUUCCUCAUCACCGAGUUCCCCCAGGGCAUCUUCGCGCUGUUGAUCGCCAUCAAAGGCAAAGACUUGUUCGUCCGAUGCUACUUGCUGUACG ........((((.(((((((.(((.........))).)))))....)).))))((((((.....(((.(((((....))))).)))...(((......))).))))))............ ( -32.40, z-score = 0.29, R) >consensus CUCUUCUGCUGGUGGCGGUGCUGGUGCUGUUCCUGAUCACCGAAUUUCCGCAGGGAUUACUCGGACUGCUGUCCGGGAUGAUGGAGAAGUGCUUCUUCGCCCACUGCUAUCCGCCGUUCG .......((.((((((((((..(((.....((((((.((.......((((.((......))))))....)).))))))....((((((....))))))))))))))))))).))...... (-20.93 = -21.41 + 0.48)

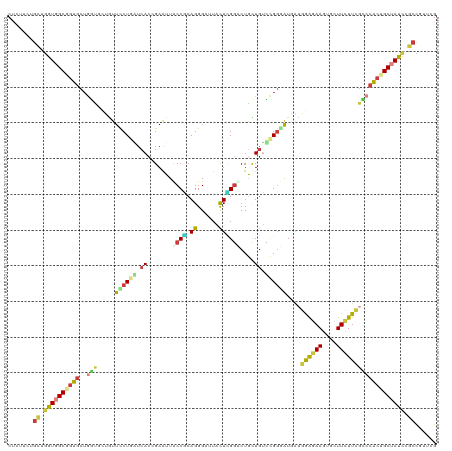
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:35 2011