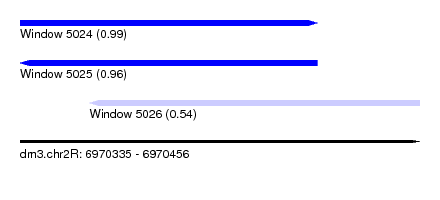
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,970,335 – 6,970,456 |
| Length | 121 |
| Max. P | 0.994399 |

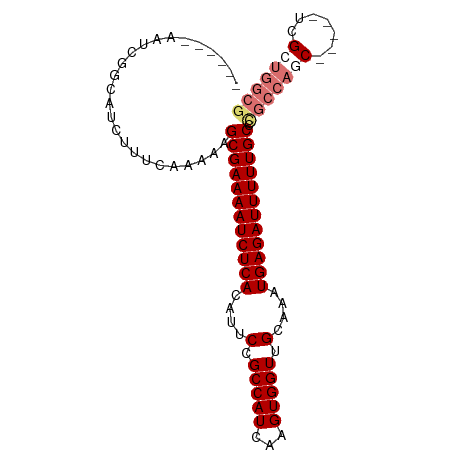
| Location | 6,970,335 – 6,970,425 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Shannon entropy | 0.23800 |
| G+C content | 0.46681 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -23.08 |
| Energy contribution | -24.70 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6970335 90 + 21146708 ------AAUCGGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCUGCCACCACCUCUCGCUGGCG ------...................((((((((((((....(.(((((...))))).)....))))))))))))..((((.(.......).)))). ( -23.90, z-score = -2.18, R) >droYak2.chr2R 18966198 87 - 21139217 AAUCGUUCGUGGAAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCUGCCAGCGG---ACG------ ...(((((((((.............((((((((((((....(.(((((...))))).)....))))))))))))...))).)))---)))------ ( -26.69, z-score = -2.65, R) >droSec1.super_1 4537564 85 + 14215200 ------AAUCGGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAGC-----UUGCUGGCG ------...................((((((((((((....(.(((((...))))).)....)))))))))))).((((((.-----...)))))) ( -29.30, z-score = -3.37, R) >droSim1.chr2R 5511311 85 + 19596830 ------AAUCGGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAGC-----UUGCUGGCG ------...................((((((((((((....(.(((((...))))).)....)))))))))))).((((((.-----...)))))) ( -29.30, z-score = -3.37, R) >consensus ______AAUCGGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAGC_____UCGCUGGCG .........................((((((((((((....(.(((((...))))).)....)))))))))))).(((((((.......))))))) (-23.08 = -24.70 + 1.62)

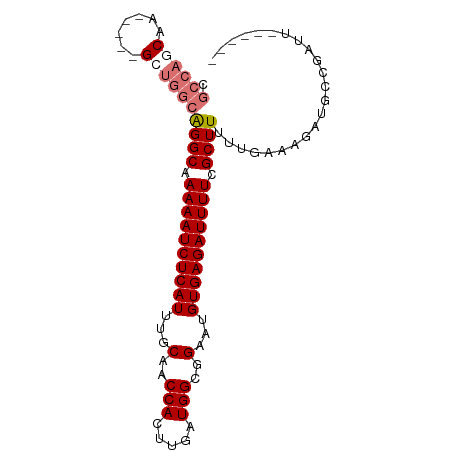
| Location | 6,970,335 – 6,970,425 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Shannon entropy | 0.23800 |
| G+C content | 0.46681 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -23.55 |
| Energy contribution | -24.80 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6970335 90 - 21146708 CGCCAGCGAGAGGUGGUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCCGAUU------ .((((.(....).))))(((((((.((((((((((...(..(((.....)))..)...)))))))))).)))..((....))))))....------ ( -31.50, z-score = -2.40, R) >droYak2.chr2R 18966198 87 + 21139217 ------CGU---CCGCUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUUCCACGAACGAUU ------(((---.((.(((.((((.((((((((((...(..(((.....)))..)...)))))))))).)))).((....))..))))).)))... ( -24.30, z-score = -1.45, R) >droSec1.super_1 4537564 85 - 14215200 CGCCAGCAA-----GCUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCCGAUU------ ((((((...-----.))))))((((((((((((((...(..(((.....)))..)...))))))))))..(((.....))).))))....------ ( -30.70, z-score = -2.68, R) >droSim1.chr2R 5511311 85 - 19596830 CGCCAGCAA-----GCUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCCGAUU------ ((((((...-----.))))))((((((((((((((...(..(((.....)))..)...))))))))))..(((.....))).))))....------ ( -30.70, z-score = -2.68, R) >consensus CGCCAGCAA_____GCUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCCGAUU______ .((((((.......))))))((((.((((((((((...(..(((.....)))..)...)))))))))).))))....................... (-23.55 = -24.80 + 1.25)

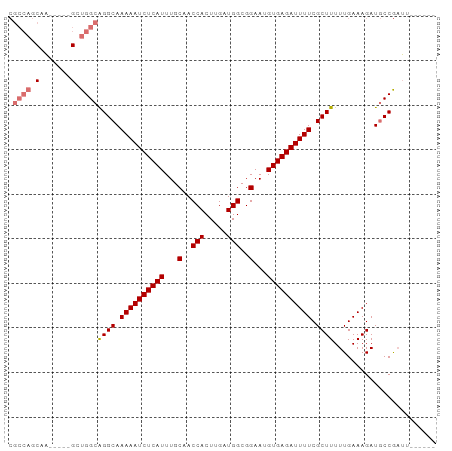
| Location | 6,970,356 – 6,970,456 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.81 |
| Shannon entropy | 0.51936 |
| G+C content | 0.47925 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6970356 100 - 21146708 -------CAUUUUCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCGAGAGGUGGUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUC -------(((((((((((...)))).))).)))).((.(((((.(....).))))))).....((((((((((...(..(((.....)))..)...)))))))))). ( -29.40, z-score = -1.19, R) >dp4.chr3 1002063 78 + 19779522 -------CAUUUUCCCCCCUUGACUGCUGUCGUACACGUGUC----------AGUGCAGGAUUCAGUGCUCGCUGG---------GCUGGCGGCUUAUGAAAUC--- -------.....(((......(((....)))((((.......----------.)))).)))((((..((.((((..---------...))))))...))))...--- ( -19.40, z-score = 0.87, R) >droYak2.chr2R 18966225 98 + 21139217 CAUUUUACGUGUUCCGUGUUUUACGAGAGCAAUGAGUCCGUCC---------GCUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUC .(((((((((((((((((...)))).))))......((((((.---------..(((...(((((.......)))))..)))......))))))))))))))).... ( -26.10, z-score = -0.72, R) >droSec1.super_1 4537585 95 - 14215200 -------AAUUUUCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCAA-----GCUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUC -------.....((..(((((.....)))))..))(((((((((...-----.))))))))).((((((((((...(..(((.....)))..)...)))))))))). ( -32.90, z-score = -2.88, R) >droSim1.chr2R 5511332 95 - 19596830 -------CAUUUUCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCAA-----GCUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUC -------(((((((((((...)))).))).)))).(((((((((...-----.))))))))).((((((((((...(..(((.....)))..)...)))))))))). ( -34.90, z-score = -3.51, R) >consensus _______CAUUUUCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGC_A_____GCUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUC .......(((.((((((...........((((((((...(((.((...........)).)))......))))).)))............)))))).)))........ (-12.66 = -13.06 + 0.40)

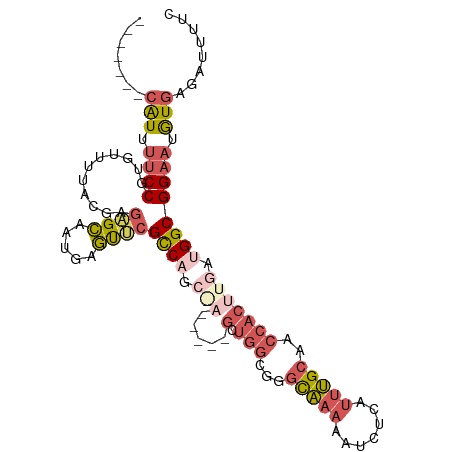
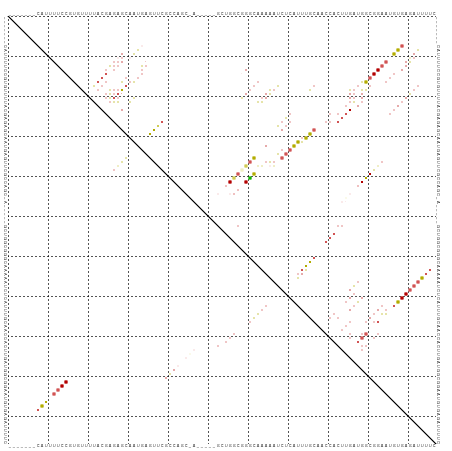
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:26 2011