| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,961,079 – 6,961,170 |
| Length | 91 |
| Max. P | 0.630977 |

| Location | 6,961,079 – 6,961,170 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 71.90 |
| Shannon entropy | 0.60273 |
| G+C content | 0.36081 |
| Mean single sequence MFE | -14.29 |
| Consensus MFE | -6.28 |
| Energy contribution | -6.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
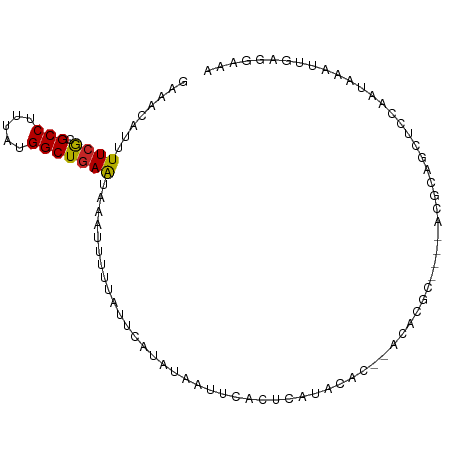
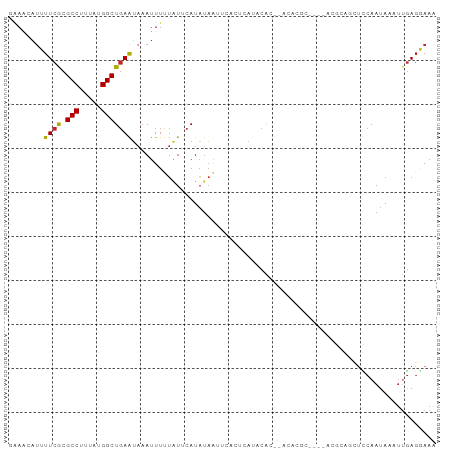
>dm3.chr2R 6961079 91 + 21146708 GGAACAUUUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACAC--ACACGC----ACGCAUCUCCAAUAAAUUGAGGAAA ...........(((((.....)))(((((((....)))))))..................--....))----.....((..(((....)))..)).. ( -13.30, z-score = -1.52, R) >droSim1.chr2R 5501664 91 + 19596830 GGAACAUUUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACAC--ACACGC----ACGCAGUUUCAAUAAAUUGAGGAAA ......((((((((((.....)))(((((((....)))))))..................--....))----...((((((....)))))).))))) ( -14.30, z-score = -1.64, R) >droSec1.super_1 4528361 91 + 14215200 GGAACAUUUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACAC--ACACGC----ACGCAGUUCCAAUAAAUUGAGGAAA (((((......(((((.....)))(((((((....)))))))..................--....))----.....)))))............... ( -16.40, z-score = -2.30, R) >droYak2.chr2R 18956792 93 - 21139217 AAAACAUUUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACACACACACGC----ACGCAGUUCCAAUAAAUUGAGGAAA ......((((((((((.....)))(((((((....)))))))........................))----...(((((......))))).))))) ( -13.80, z-score = -1.71, R) >droEre2.scaffold_4845 3753697 90 - 22589142 -------UUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACACACACGCACUUGUACGCAGUUCCAAUAAAUUGAGGAAA -------.((((((((.....)))(((((((....)))))))......................)).......(.(((((......))))).)))). ( -13.60, z-score = -1.06, R) >droAna3.scaffold_13266 669109 93 + 19884421 CGAGCAUUUUCGCGCCAUUAUGGCUGAAUAAAUUUUUAUUCAUACAAUUCAGUCAUACACCCACACGC----ACGCAGUUUCAAUAAAUUGAGGAAA ......((((((((....(((((((((((.................)))))))))))........)))----...((((((....)))))).))))) ( -16.23, z-score = -1.33, R) >droWil1.scaffold_180697 3286305 91 + 4168966 UAAGCAUUUUCGCGCCUUUAUGGCUCAGGCAAUUUCAAUAAAUUUCGAAC-CCCCCGCGC-----CCCUCUUACAAUUCCUCAAUAAGCAAACGAAG ((((......((((((.....)))...((....(((..........))).-...))))).-----....))))........................ ( -9.50, z-score = 0.92, R) >droVir3.scaffold_12823 2228693 89 - 2474545 GAAAAAUGUUCACGCCUUUAUGGCUGAAUAAAUUUCUAUACAUAUAA-------AGUCGCGUGCGUUUGC-UACAAGCCUCCAAUAAUUUGAGCAAA ((((..((((((.(((.....)))))))))..))))...........-------...........(((((-(.((((..........)))))))))) ( -16.70, z-score = -0.94, R) >droMoj3.scaffold_6496 26777460 89 - 26866924 GAAAAAUGUUCACGCCUUUAUGGCUGAAUAAAUUUUUAUACAUAUAA---AGUCGGGCGC--GCUCGGU---ACAAACCUUCAAUAAUUCGAGCAAA ......((((((.(((.....))))))))).................---..........--((((((.---................))))))... ( -17.63, z-score = -1.13, R) >droGri2.scaffold_15245 6400829 79 + 18325388 GAAAAAUGUUCACGCCCCUAUGGCUGAAUAAAUUUGUGUACAUAUAA-------AAACGC-----------UGAAAGCCAUCAAUAAUUUGAGAAAA ......((((((.(((.....))))))))).....((((........-------..))))-----------.........((((....))))..... ( -11.40, z-score = -1.19, R) >consensus GAAACAUUUUCGCGCCUUUAUGGCUGAAUAAAUUUUUAUUCAUAUAAUUCACUCAUACAC__ACACGC____ACGCAGCUCCAAUAAAUUGAGGAAA ........((((.(((.....)))))))..................................................................... ( -6.28 = -6.08 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:21 2011