
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,530,513 – 2,530,584 |
| Length | 71 |
| Max. P | 0.996739 |

| Location | 2,530,513 – 2,530,584 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 65.39 |
| Shannon entropy | 0.60513 |
| G+C content | 0.53320 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
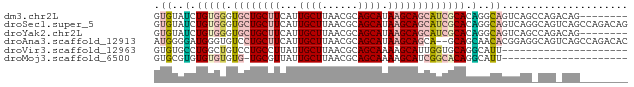
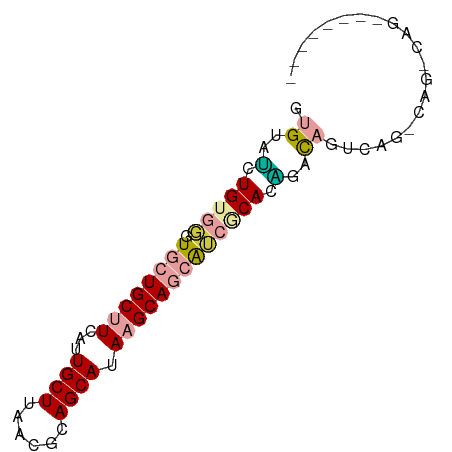
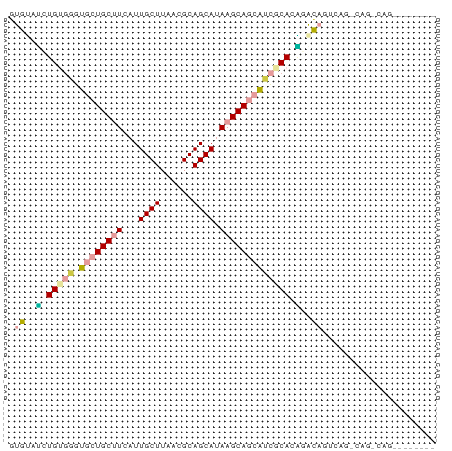
>dm3.chr2L 2530513 71 + 23011544 GUGUAUCUGUGGGUGCUGCUUCAUUGCUUAACGCAGCAUAAGCAGCAUCGCACAGGCAGUCAGCCAGACAG-------- .....((((((((((((((((...((((......)))).)))))))))).))))))..(((.....)))..-------- ( -30.10, z-score = -3.17, R) >droSec1.super_5 704388 79 + 5866729 GUGUAUCUGUGGGUGCUGCUUCAUUGCUUAACGCAGCAUAAGCAGCAUCGCACAGGCAGUCAGGCAGUCAGCCAGACAG .....((((((((((((((((...((((......)))).)))))))))).))))))..(((.(((.....))).))).. ( -35.70, z-score = -4.14, R) >droYak2.chr2L 2522974 71 + 22324452 GUGUAUCUGUGGGUGCUGCUUCAUUGCUUAACGCAGCAUAAGCAGCAUCGCACAGGCAGUCAGCCAGACAG-------- .....((((((((((((((((...((((......)))).)))))))))).))))))..(((.....)))..-------- ( -30.10, z-score = -3.17, R) >droAna3.scaffold_12913 295144 77 - 441482 AUGGGGAUGGGUGUCCUGCUUCAUUGCUUAACGCAGCAUAAGCAGCA--GCAGCAACACGGAGGCAGUCAGCCAGACAC ....(..(((.((..(((((((.(((((....((.((....)).)).--..)))))....))))))).)).)))..).. ( -26.10, z-score = -1.45, R) >droVir3.scaffold_12963 9973386 58 + 20206255 GUGUGCCUGGCUGUCCUGCCUUAUUGCUUAACGCAGCAAAAGCAUUGGUGCAGGCAUU--------------------- ..(((((((((......)))............(((.(((.....))).))))))))).--------------------- ( -18.70, z-score = -0.18, R) >droMoj3.scaffold_6500 31034254 57 - 32352404 GUGCGUGUGUGUGUG-UGCGUUAUUGCUUAACGCAGCAAAAGCAUCGGCACAGGCAUU--------------------- ((((.(((((.....-(((((((.....)))))))((....))....))))).)))).--------------------- ( -21.90, z-score = -1.33, R) >consensus GUGUAUCUGUGGGUGCUGCUUCAUUGCUUAACGCAGCAUAAGCAGCAUCGCACAGACAGUCAG_CAG_CAG________ .((..(.(((((.((((((((...((((......)))).))))))))))))).)..))..................... (-13.03 = -13.78 + 0.75)
| Location | 2,530,513 – 2,530,584 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 65.39 |
| Shannon entropy | 0.60513 |
| G+C content | 0.53320 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -11.58 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
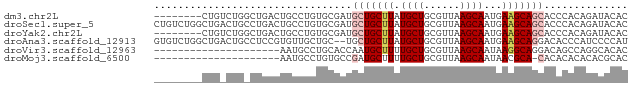
>dm3.chr2L 2530513 71 - 23011544 --------CUGUCUGGCUGACUGCCUGUGCGAUGCUGCUUAUGCUGCGUUAAGCAAUGAAGCAGCACCCACAGAUACAC --------.((((((((.....))).(((.(.((((((((.((((......))))...)))))))).)))))))))... ( -28.30, z-score = -3.22, R) >droSec1.super_5 704388 79 - 5866729 CUGUCUGGCUGACUGCCUGACUGCCUGUGCGAUGCUGCUUAUGCUGCGUUAAGCAAUGAAGCAGCACCCACAGAUACAC ..(((.(((.....))).)))...(((((.(.((((((((.((((......))))...)))))))).))))))...... ( -33.50, z-score = -4.51, R) >droYak2.chr2L 2522974 71 - 22324452 --------CUGUCUGGCUGACUGCCUGUGCGAUGCUGCUUAUGCUGCGUUAAGCAAUGAAGCAGCACCCACAGAUACAC --------.((((((((.....))).(((.(.((((((((.((((......))))...)))))))).)))))))))... ( -28.30, z-score = -3.22, R) >droAna3.scaffold_12913 295144 77 + 441482 GUGUCUGGCUGACUGCCUCCGUGUUGCUGC--UGCUGCUUAUGCUGCGUUAAGCAAUGAAGCAGGACACCCAUCCCCAU (((((((((.....))).....((((((..--(((.((....)).)))...))))))......)))))).......... ( -23.90, z-score = -1.09, R) >droVir3.scaffold_12963 9973386 58 - 20206255 ---------------------AAUGCCUGCACCAAUGCUUUUGCUGCGUUAAGCAAUAAGGCAGGACAGCCAGGCACAC ---------------------..(((((((.....((((((((((......))))..)))))).....).))))))... ( -19.70, z-score = -1.45, R) >droMoj3.scaffold_6500 31034254 57 + 32352404 ---------------------AAUGCCUGUGCCGAUGCUUUUGCUGCGUUAAGCAAUAACGCA-CACACACACACGCAC ---------------------..(((.((((.....((....))(((((((.....)))))))-......)))).))). ( -15.50, z-score = -1.26, R) >consensus ________CUG_CUG_CUGACUGCCUCUGCGAUGCUGCUUAUGCUGCGUUAAGCAAUGAAGCAGCACACACAGACACAC .................................(((((((.((((......))))...))))))).............. (-11.58 = -12.30 + 0.72)
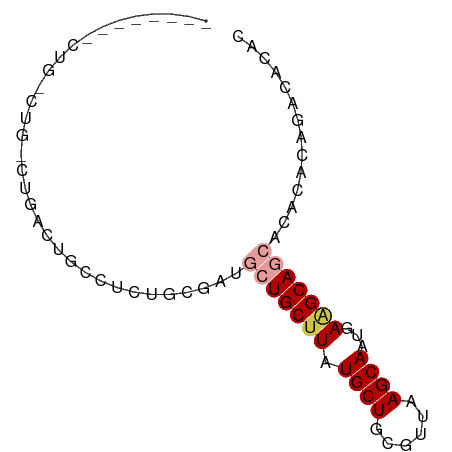
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:37 2011