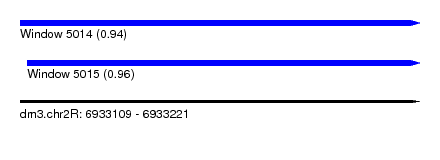
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,933,109 – 6,933,221 |
| Length | 112 |
| Max. P | 0.964798 |

| Location | 6,933,109 – 6,933,221 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.37299 |
| G+C content | 0.36489 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
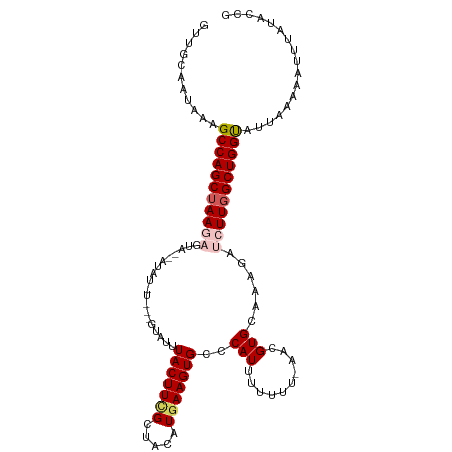
>dm3.chr2R 6933109 112 + 21146708 GUUGCAAUAAAGCCAGCUAAGAGUAAUAUAUAUGUGGUACUUACUUCGCUACAUGAAGUGCCCAUUUUUUUUAACGUGCAAAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG .......(((.(((((((((((.(((.....(((.(((((((.(..........))))))))))).....)))...........))))))))))).)))............. ( -26.60, z-score = -1.83, R) >droEre2.scaffold_4845 3726970 109 - 22589142 GUUGCCAUAAAGCCAGCUAAGAGUCGAAUAUUUGUGGUAUUUACUUCGAAACUUAAAGUGCUCAUUUUUUU-AGAGUGCAAAGACGUUGGCUGGC-UU-AAAAUGUAUAGCG ((((.(((.(((((((((((..(((.....((((.(((....))).)))).......(..(((........-.)))..)...))).)))))))))-))-...)))..)))). ( -31.90, z-score = -2.73, R) >droYak2.chr2R 18928510 109 - 21139217 GUGGCAAUAAAGCCAGCAAAGAGCUGAAUAUGUCAAGUAUUUACUUUGAUACUUGAAGUGAUCAUUUUUU--AGAGUGCAACGAUGUUGGCUGGC-UUUAAAAUUUAUACCG .......((((((((((((((((.(((.(((.(((((((((......))))))))).))).)))))))))--......((((...))))))))))-))))............ ( -32.90, z-score = -3.63, R) >droSec1.super_1 4500512 97 + 14215200 UUUGCAAUAAAGCCAGCUAAGAGUA--------------UUUACUUCGCUACAUGAAGUGCCCAUUUUUUU-AACGUGCACAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG .......(((.(((((((((((...--------------.....((((.....))))((((.(.((.....-)).).))))...))))))))))).)))............. ( -22.70, z-score = -2.13, R) >droSim1.chr2R 5473600 97 + 19596830 GUUGCAAUAAAGCCAGCUAAGUGUA--------------UUUACUUCGCUACAUGAAGUGCCCAUUUUUUU-AACGUGCACAGAUCUUGGCUGGUAUUAAAAAUUUGUACCG (.((((((((.(((((((((((((.--------------...((((((.....))))))((.(.((.....-)).).)))))...)))))))))).))).....))))).). ( -22.10, z-score = -1.15, R) >consensus GUUGCAAUAAAGCCAGCUAAGAGUA__AUAU_U___GUAUUUACUUCGCUACAUGAAGUGCCCAUUUUUUU_AACGUGCAAAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG ...........(((((((((((...................(((((((.....)))))))..(((.((....)).)))......)))))))))))................. (-15.98 = -16.58 + 0.60)

| Location | 6,933,111 – 6,933,221 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.41 |
| Shannon entropy | 0.37320 |
| G+C content | 0.36436 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
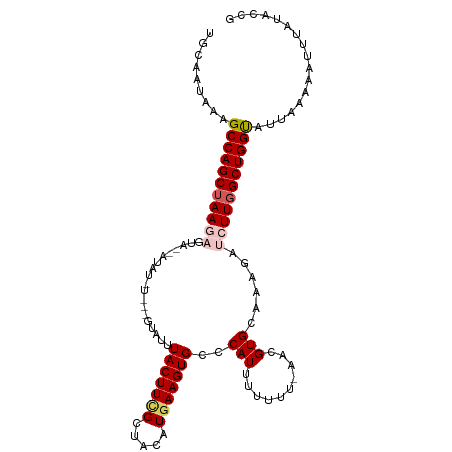
>dm3.chr2R 6933111 110 + 21146708 UGCAAUAAAGCCAGCUAAGAGUAAUAUAUAUGUGGUACUUACUUCGCUACAUGAAGUGCCCAUUUUUUUUAACGUGCAAAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG .....(((.(((((((((((.(((.....(((.(((((((.(..........))))))))))).....)))...........))))))))))).)))............. ( -26.60, z-score = -2.06, R) >droEre2.scaffold_4845 3726972 107 - 22589142 UGCCAUAAAGCCAGCUAAGAGUCGAAUAUUUGUGGUAUUUACUUCGAAACUUAAAGUGCUCAUUUUUUU-AGAGUGCAAAGACGUUGGCUGGC-UU-AAAAUGUAUAGCG .((.((((((((((((((..(((.....((((.(((....))).)))).......(..(((........-.)))..)...))).)))))))))-))-......))).)). ( -31.30, z-score = -2.78, R) >droYak2.chr2R 18928512 107 - 21139217 GGCAAUAAAGCCAGCAAAGAGCUGAAUAUGUCAAGUAUUUACUUUGAUACUUGAAGUGAUCAUUUUUU--AGAGUGCAACGAUGUUGGCUGGC-UUUAAAAUUUAUACCG .....((((((((((((((((.(((.(((.(((((((((......))))))))).))).)))))))))--......((((...))))))))))-))))............ ( -32.90, z-score = -4.00, R) >droSec1.super_1 4500514 95 + 14215200 UGCAAUAAAGCCAGCUAAGAGUA--------------UUUACUUCGCUACAUGAAGUGCCCAUUUUUUU-AACGUGCACAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG .....(((.(((((((((((...--------------.....((((.....))))((((.(.((.....-)).).))))...))))))))))).)))............. ( -22.70, z-score = -2.19, R) >droSim1.chr2R 5473602 95 + 19596830 UGCAAUAAAGCCAGCUAAGUGUA--------------UUUACUUCGCUACAUGAAGUGCCCAUUUUUUU-AACGUGCACAGAUCUUGGCUGGUAUUAAAAAUUUGUACCG ((((((((.(((((((((((((.--------------...((((((.....))))))((.(.((.....-)).).)))))...)))))))))).))).....)))))... ( -21.50, z-score = -1.20, R) >consensus UGCAAUAAAGCCAGCUAAGAGUA__AUAU_U___GUAUUUACUUCGCUACAUGAAGUGCCCAUUUUUUU_AACGUGCAAAGAUCUUGGCUGGUAUUAAAAAUUUAUACCG .........(((((((((((...................(((((((.....)))))))..(((.((....)).)))......)))))))))))................. (-15.98 = -16.58 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:17 2011