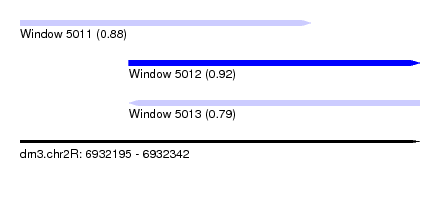
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,932,195 – 6,932,342 |
| Length | 147 |
| Max. P | 0.918768 |

| Location | 6,932,195 – 6,932,302 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Shannon entropy | 0.13182 |
| G+C content | 0.34370 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
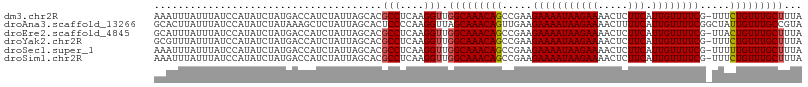
>dm3.chr2R 6932195 107 + 21146708 AAAUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUA ......................................(((....))).(((((((((.....(((((((((((.....))).)))))))).-...)))))))))... ( -20.50, z-score = -1.82, R) >droAna3.scaffold_13266 638453 108 + 19884421 GCACUUAUUUAUCCAUAUCUAUAAAGCUCUAUUAGCACUCCCCAAGGUUAGCAAACAGUUGAAGAAAAUAAGAAAACUUUUCAUUGUUUUCGGCUAUUGUUUGCCGUA ((.......................(((.....)))..............(((((((((.(..(((((((((((.....))).))))))))..).))))))))).)). ( -20.60, z-score = -1.52, R) >droEre2.scaffold_4845 3726039 107 - 22589142 GCAUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUACUGUUUGCUUUA ......................................(((....))).(((((((((.....(((((((((((.....))).)))))))).-...)))))))))... ( -20.50, z-score = -1.54, R) >droYak2.chr2R 18927575 107 - 21139217 GCGUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUA ((...........(((....)))...........))..(((....))).(((((((((.....(((((((((((.....))).)))))))).-...)))))))))... ( -21.45, z-score = -1.63, R) >droSec1.super_1 4499592 107 + 14215200 AAAUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUUUGUUUGCUUUA .................................((((.((.....(((((.....)))))...(((((((((((.....))).)))))))).-.....)).))))... ( -18.40, z-score = -1.07, R) >droSim1.chr2R 5472709 107 + 19596830 AAAUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUA ......................................(((....))).(((((((((.....(((((((((((.....))).)))))))).-...)))))))))... ( -20.50, z-score = -1.82, R) >consensus AAAUUUAUUUAUCCAUAUCUAUGACCAUCUAUUAGCACGCCUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG_UUUCUGUUUGCUUUA ......................................(((....))).(((((((((.....(((((((((((.....))).)))))))).....)))))))))... (-19.02 = -19.13 + 0.11)

| Location | 6,932,235 – 6,932,342 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Shannon entropy | 0.21398 |
| G+C content | 0.39879 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -24.53 |
| Energy contribution | -24.06 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
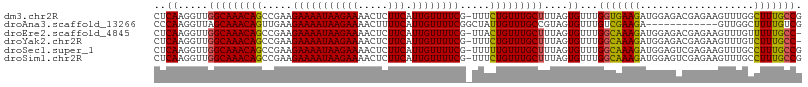
>dm3.chr2R 6932235 107 + 21146708 CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUAGUGUUUGGUGAAGAUGGAGACGAGAAGUUUGGCUUUGCCG .........(((((..(((((((((............)))))....((((((-((((((((((((.........)))..)))))))))))))))....))))))))). ( -32.00, z-score = -2.04, R) >droAna3.scaffold_13266 638493 96 + 19884421 CCCAAGGUUAGCAAACAGUUGAAGAAAAUAAGAAAACUUUUCAUUGUUUUCGGCUAUUGUUUGCCGUAGUGUUUGUCGAAGA------------GUUGGCUUUUGUCG .((((..(..(((((((......(((((((((((.....))).))))))))(((........)))....)))))))..)...------------.))))......... ( -22.50, z-score = -1.31, R) >droEre2.scaffold_4845 3726079 106 - 22589142 CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUACUGUUUGCUUUAGUGUUUGGCAAAGAUGGAGACGAGAAGUUUGUUUUUGCC- ..((.....(((((((((.....(((((((((((.....))).)))))))).-...)))))))))....))...(((((((((..((((.....)))))))))))))- ( -30.00, z-score = -2.00, R) >droYak2.chr2R 18927615 106 - 21139217 CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUAGUGUUUGGCAAAGAUGGAGACGAGAAGUUUGUCUUUGCC- ..((.....(((((((((.....(((((((((((.....))).)))))))).-...)))))))))....))...(((((((((..((((.....)))))))))))))- ( -32.60, z-score = -2.53, R) >droSec1.super_1 4499632 107 + 14215200 CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUUUGUUUGCUUUAGUGUUUGGCAAAGAUGGAGUCGAGAAGUUUGCCUUUGCCG .....(((.(((((((....(((((............))))).(((.(((((-((((((((.((......))..))))))))))))).)))...)))))))...))). ( -33.00, z-score = -2.57, R) >droSim1.chr2R 5472749 107 + 19596830 CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG-UUUCUGUUUGCUUUAGUGUUUGGCAAAGAUGGAGUCGAGAAGUUUGCCUUUGCCG .....(((.(((((((....(((((............)))))....((((((-(((((.((((((.........)))))).).)))).)))))))))))))...))). ( -32.10, z-score = -2.18, R) >consensus CUCAAGGUUGGCAAACAGCCGAAGAAAAUAAGAAAACUCUUCAUUGUUUUCG_UUUCUGUUUGCUUUAGUGUUUGGCAAAGAUGGAGACGAGAAGUUUGCCUUUGCCG ..((.....(((((((((.....(((((((((((.....))).)))))))).....)))))))))....))...(((((((...................))))))). (-24.53 = -24.06 + -0.47)
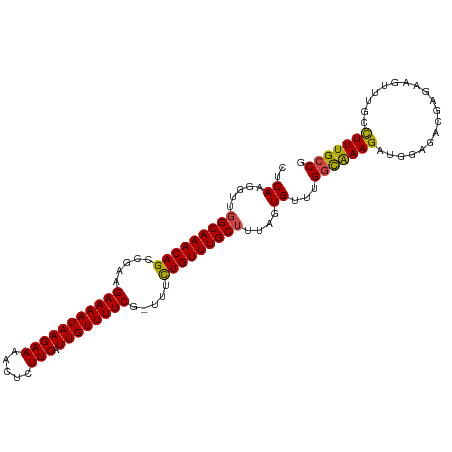
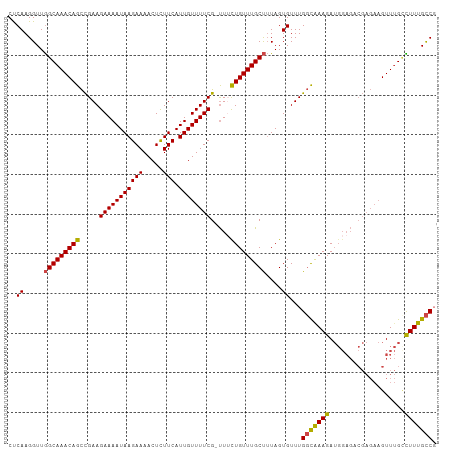
| Location | 6,932,235 – 6,932,342 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Shannon entropy | 0.21398 |
| G+C content | 0.39879 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.60 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6932235 107 - 21146708 CGGCAAAGCCAAACUUCUCGUCUCCAUCUUCACCAAACACUAAAGCAAACAGAAA-CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG .(((((((((.......((((.((...........................)).)-))).......((((((..........))))))))))..)))))......... ( -20.33, z-score = -1.38, R) >droAna3.scaffold_13266 638493 96 - 19884421 CGACAAAAGCCAAC------------UCUUCGACAAACACUACGGCAAACAAUAGCCGAAAACAAUGAAAAGUUUUCUUAUUUUCUUCAACUGUUUGCUAACCUUGGG .........((((.------------.....(.((((((....(((........)))((((((........))))))..............))))))).....)))). ( -16.60, z-score = -1.62, R) >droEre2.scaffold_4845 3726079 106 + 22589142 -GGCAAAAACAAACUUCUCGUCUCCAUCUUUGCCAAACACUAAAGCAAACAGUAA-CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG -((((((.....................))))))..........(((((((((..-((((.......(((......)))......))))))))))))).......... ( -21.22, z-score = -1.88, R) >droYak2.chr2R 18927615 106 + 21139217 -GGCAAAGACAAACUUCUCGUCUCCAUCUUUGCCAAACACUAAAGCAAACAGAAA-CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG -((((((((...((.....)).....))))))))..........((((((((...-((((.......(((......)))......)))).)))))))).......... ( -26.12, z-score = -2.90, R) >droSec1.super_1 4499632 107 - 14215200 CGGCAAAGGCAAACUUCUCGACUCCAUCUUUGCCAAACACUAAAGCAAACAAAAA-CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG .((((((((.................))))))))..........(((((((....-.((((((........)))))).......(....).))))))).......... ( -22.03, z-score = -1.49, R) >droSim1.chr2R 5472749 107 - 19596830 CGGCAAAGGCAAACUUCUCGACUCCAUCUUUGCCAAACACUAAAGCAAACAGAAA-CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG .((((((((.................))))))))..........((((((((...-((((.......(((......)))......)))).)))))))).......... ( -25.25, z-score = -2.11, R) >consensus CGGCAAAGGCAAACUUCUCGUCUCCAUCUUUGCCAAACACUAAAGCAAACAGAAA_CGAAAACAAUGAAGAGUUUUCUUAUUUUCUUCGGCUGUUUGCCAACCUUGAG .((((((((.................))))))))..........((((((((.....((((((........)))))).......(....))))))))).......... (-17.10 = -18.60 + 1.50)
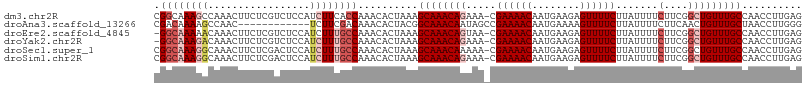
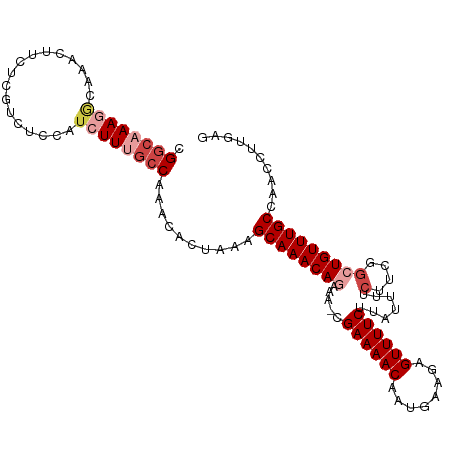

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:16 2011