
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,931,184 – 6,931,281 |
| Length | 97 |
| Max. P | 0.965940 |

| Location | 6,931,184 – 6,931,281 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.17495 |
| G+C content | 0.44849 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.75 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 6931184 97 + 21146708 CCACUCAAGGCUAUAUGGCCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCC--------UCCGUAUCCUGCAGUAUAUUCAAAUGUGUGUGUGUG-UGUG--- ...(((..((((((((....))))))))))).((((((..((((.((((........--------..))))))))(((.(((((......)))))))))))-))).--- ( -25.30, z-score = -0.89, R) >droSim1.chr2R 5471455 101 + 19596830 CCACUCAAGGCUAUAUGGCCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCC--------UCCGUAUCCUGCAGUAUAUUCAAAUGUGUGUGUGUGGUGUGUGG ((((....((((((((....)))))))).((((.(((....((((.....))))...--------..))).))))(((.(((((.((......)).))))).))))))) ( -27.80, z-score = -0.62, R) >droSec1.super_1 4498579 97 + 14215200 CCACUCAAGGCUAUAUGGCCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCC--------UCCGUAUCCUGCAGUAUAUUCAAAUGUGUGUGUGUG-UGUG--- ...(((..((((((((....))))))))))).((((((..((((.((((........--------..))))))))(((.(((((......)))))))))))-))).--- ( -25.30, z-score = -0.89, R) >droYak2.chr2R 18926496 104 - 21139217 CCACUCAAGGCUAUAUAGUCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCCCCAUAGUGCCCAUAUCCUGCAGUAUAUUUAAAUGUUUGUGUGUG-CGU---- ........((((((((....))))))))..(.(((((((((((((.....)))).....(((.(((.........))).))).........))))))))).-)..---- ( -24.80, z-score = -1.15, R) >droEre2.scaffold_4845 3725051 94 - 22589142 CCACUCAAGGCUAUAUAGUCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCC--------CCCAUAUCCUGCAGUACAUUCAAAUGCUUGUGUGUG-C------ ...(((..((((((((....))))))))))).((((((((((((.((((........--------..))))))))(((...........))))))))))).-.------ ( -24.90, z-score = -1.84, R) >consensus CCACUCAAGGCUAUAUGGCCAUAUAGCCGAGGACAUGCAAAGGAAUAUGUUUCUCCC________UCCGUAUCCUGCAGUAUAUUCAAAUGUGUGUGUGUG_UGUG___ .(((.((.((((((((....))))))))(((.((.((((..(((.((((..................)))))))))))))...))).......)).))).......... (-23.15 = -22.75 + -0.40)

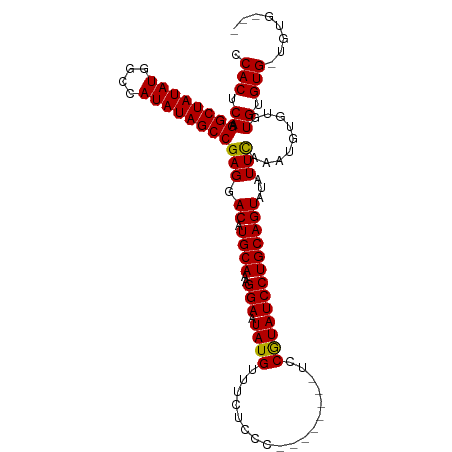
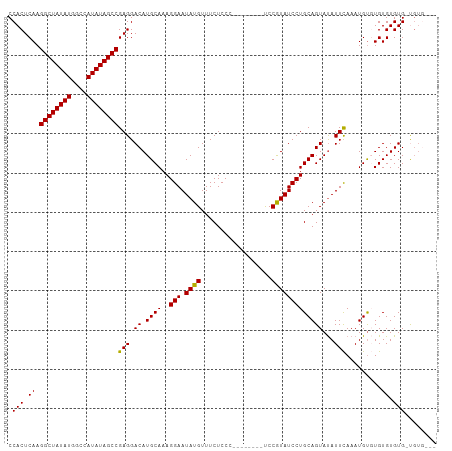
| Location | 6,931,184 – 6,931,281 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.17495 |
| G+C content | 0.44849 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.44 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6931184 97 - 21146708 ---CACA-CACACACACACAUUUGAAUAUACUGCAGGAUACGGA--------GGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGGCCAUAUAGCCUUGAGUGG ---....-..........((((..(.......(.((((((((((--------((((........))))))))..)))))))((((((((....)))))))))..)))). ( -29.40, z-score = -3.26, R) >droSim1.chr2R 5471455 101 - 19596830 CCACACACCACACACACACAUUUGAAUAUACUGCAGGAUACGGA--------GGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGGCCAUAUAGCCUUGAGUGG .......((((.((..................(.((((((((((--------((((........))))))))..)))))))((((((((....)))))))).)).)))) ( -30.20, z-score = -3.14, R) >droSec1.super_1 4498579 97 - 14215200 ---CACA-CACACACACACAUUUGAAUAUACUGCAGGAUACGGA--------GGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGGCCAUAUAGCCUUGAGUGG ---....-..........((((..(.......(.((((((((((--------((((........))))))))..)))))))((((((((....)))))))))..)))). ( -29.40, z-score = -3.26, R) >droYak2.chr2R 18926496 104 + 21139217 ----ACG-CACACACAAACAUUUAAAUAUACUGCAGGAUAUGGGCACUAUGGGGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGACUAUAUAGCCUUGAGUGG ----...-..........(((((((.......(.((((((((.(......((((((........))))))).)))))))))((((((((....))))))))))))))). ( -28.10, z-score = -2.20, R) >droEre2.scaffold_4845 3725051 94 + 22589142 ------G-CACACACAAGCAUUUGAAUGUACUGCAGGAUAUGGG--------GGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGACUAUAUAGCCUUGAGUGG ------.-..(((.(((((((....))))...(.((((((((.(--------(((.(((.....))))))).)))))))))((((((((....))))))))))).))). ( -30.00, z-score = -2.48, R) >consensus ___CACA_CACACACACACAUUUGAAUAUACUGCAGGAUACGGA________GGGAGAAACAUAUUCCUUUGCAUGUCCUCGGCUAUAUGGCCAUAUAGCCUUGAGUGG ..................(((((((.......(.((((((.(.(........(((((.......))))).).).)))))))((((((((....))))))))))))))). (-21.84 = -21.44 + -0.40)

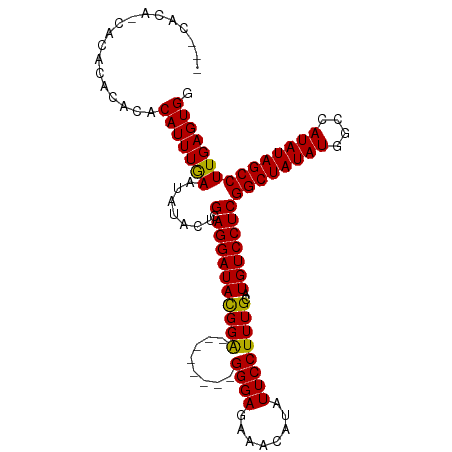
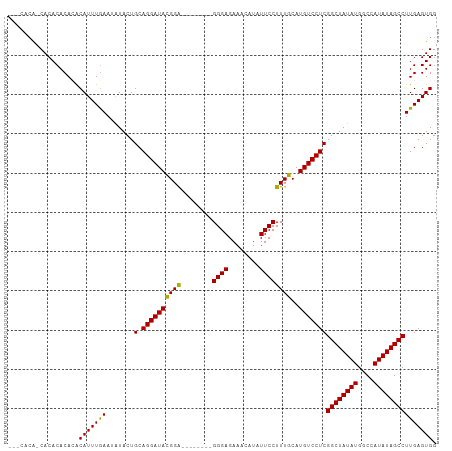
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:13 2011