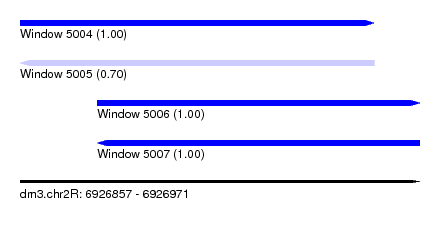
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,926,857 – 6,926,971 |
| Length | 114 |
| Max. P | 0.998348 |

| Location | 6,926,857 – 6,926,958 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Shannon entropy | 0.25632 |
| G+C content | 0.42298 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
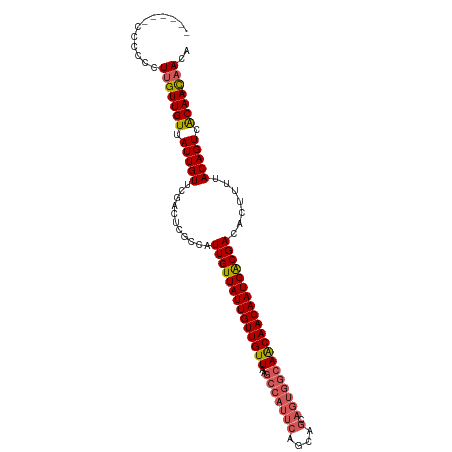
>dm3.chr2R 6926857 101 + 21146708 ------CCCCCCUUGUUGUUAUUGUUCGACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAA-CAACA ------......(((((((.(((((...........((((((((((((((..(((((((....).)))))))))))))))))))).......))))).))))-))).. ( -30.87, z-score = -4.30, R) >droSim1.chr2R 5467376 101 + 19596830 ------CCCCCCUUGUUGUUAUUGUUCGACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAA-CAACA ------......(((((((.(((((...........((((((((((((((..(((((((....).)))))))))))))))))))).......))))).))))-))).. ( -30.87, z-score = -4.30, R) >droSec1.super_1 4494288 100 + 14215200 -------CCCCCUUGUUGUUAUUGUUCGACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAA-CAACA -------.....(((((((.(((((...........((((((((((((((..(((((((....).)))))))))))))))))))).......))))).))))-))).. ( -30.87, z-score = -4.19, R) >droYak2.chr2R 18922013 100 - 21139217 -------CCCCCUUGUUGUUAUUGUUCGACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCGCAA-CAACA -------.....(((((((.(((((...........((((((((((((((..(((((((....).)))))))))))))))))))).......))))).))))-))).. ( -31.17, z-score = -4.01, R) >droAna3.scaffold_13266 633039 91 + 19884421 UUUGCACUCUCAUAGUUGUUAUUGUUCGGCCAGCUUUUGUUAUUGUUGUUAAGC-----------------AGCAACAAUGGCGACACUUUUACAGUCACAAAUAACA ..............((((((........((((((....)))(((((((((....-----------------))))))))))))(((.........)))...)))))). ( -18.90, z-score = -0.57, R) >consensus ______CCCCCCUUGUUGUUAUUGUUCGACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAA_CAACA ..............((((...((((..((((.....((((((((((((((..((((((.......)))))))))))))))))))).........)))))))).)))). (-24.48 = -24.48 + 0.00)

| Location | 6,926,857 – 6,926,958 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Shannon entropy | 0.25632 |
| G+C content | 0.42298 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.78 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6926857 101 - 21146708 UGUUG-UUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGUCGAACAAUAACAACAAGGGGGG------ (((((-((((.(((.....))).(((((((((((((((........(((....))).......))))))))))))))).....)))))))))..........------ ( -31.56, z-score = -2.32, R) >droSim1.chr2R 5467376 101 - 19596830 UGUUG-UUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGUCGAACAAUAACAACAAGGGGGG------ (((((-((((.(((.....))).(((((((((((((((........(((....))).......))))))))))))))).....)))))))))..........------ ( -31.56, z-score = -2.32, R) >droSec1.super_1 4494288 100 - 14215200 UGUUG-UUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGUCGAACAAUAACAACAAGGGGG------- (((((-((((.(((.....))).(((((((((((((((........(((....))).......))))))))))))))).....))))))))).........------- ( -31.56, z-score = -2.45, R) >droYak2.chr2R 18922013 100 + 21139217 UGUUG-UUGCGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGUCGAACAAUAACAACAAGGGGG------- (((((-(((((((..........(((((((((((((((........(((....))).......)))))))))))))))))))..)))))))).........------- ( -31.06, z-score = -2.18, R) >droAna3.scaffold_13266 633039 91 - 19884421 UGUUAUUUGUGACUGUAAAAGUGUCGCCAUUGUUGCU-----------------GCUUAACAACAAUAACAAAAGCUGGCCGAACAAUAACAACUAUGAGAGUGCAAA (((((((.(((((.........)))((((((((((.(-----------------(........)).))))))....))))...)))))))))..(((....))).... ( -15.60, z-score = 0.80, R) >consensus UGUUG_UUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGUCGAACAAUAACAACAAGGGGGG______ (((((.((((.(((.....))).(((((((((((((((........(((....))).......))))))))))))))).....)))))))))................ (-21.61 = -22.78 + 1.17)

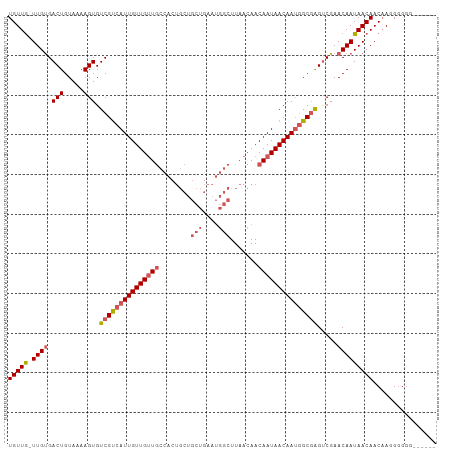
| Location | 6,926,879 – 6,926,971 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Shannon entropy | 0.17258 |
| G+C content | 0.45956 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
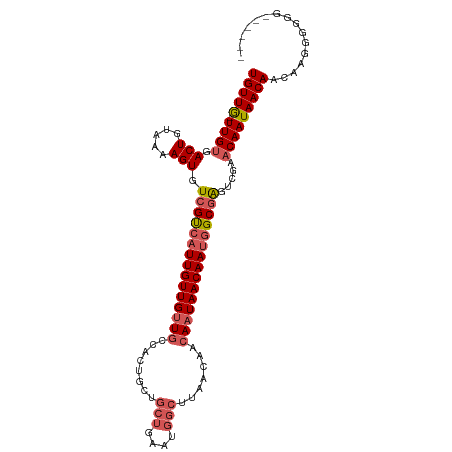
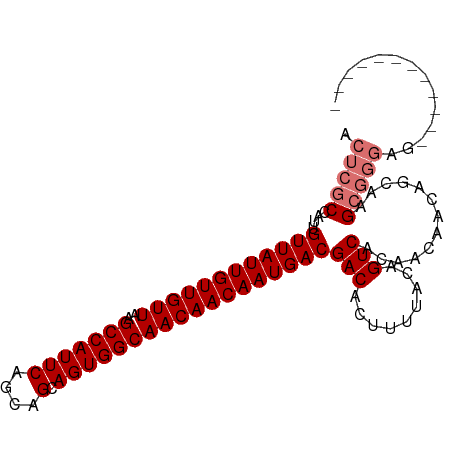
>dm3.chr2R 6926879 92 + 21146708 ACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAACAACAGCAAGCGGGAGGA---------- .(((((....((((((((((((..(((((((....).))))))))))))))))))(((.........))).............)))))....---------- ( -29.70, z-score = -3.70, R) >droSim1.chr2R 5467398 90 + 19596830 ACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAACAACAGCAAGCGGGAG------------ .(((((....((((((((((((..(((((((....).))))))))))))))))))(((.........))).............)))))..------------ ( -29.70, z-score = -4.26, R) >droSec1.super_1 4494309 90 + 14215200 ACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAACAACAGCAAGCGGGAG------------ .(((((....((((((((((((..(((((((....).))))))))))))))))))(((.........))).............)))))..------------ ( -29.70, z-score = -4.26, R) >droYak2.chr2R 18922034 102 - 21139217 ACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCGCAACAACAACGAGAACAACAGCCACCGGGAG .((((.....((((((((((((..(((((((....).))))))))))))))))))(((.........)))..........))))........((....)).. ( -27.50, z-score = -2.66, R) >consensus ACUCGCCAUUGUUAUUGUUGUUAAGCCAUUCAGCAGCAGUGGCAACAACAAUGACGACACUUUUACAGUCACAACAACAGCAAGCGGGAG____________ .(((((....((((((((((((..(((((((....).))))))))))))))))))(((.........))).............))))).............. (-25.83 = -26.82 + 1.00)

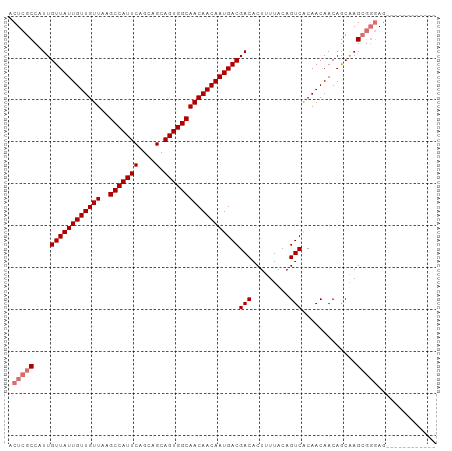
| Location | 6,926,879 – 6,926,971 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.97 |
| Shannon entropy | 0.17258 |
| G+C content | 0.45956 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.32 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
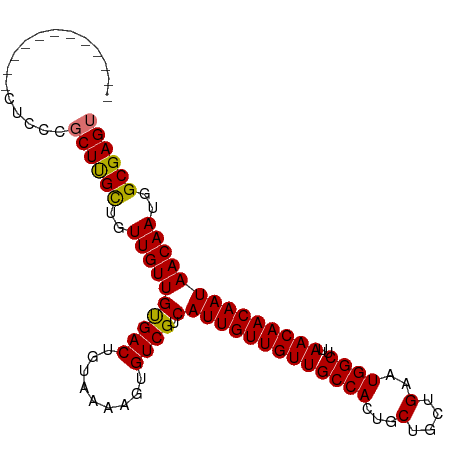
>dm3.chr2R 6926879 92 - 21146708 ----------UCCUCCCGCUUGCUGUUGUUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGU ----------.......((((((((((((((((((.........))).....(((((((((((...(....)..))))..)))))))))))))))))))))) ( -30.90, z-score = -3.65, R) >droSim1.chr2R 5467398 90 - 19596830 ------------CUCCCGCUUGCUGUUGUUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGU ------------.....((((((((((((((((((.........))).....(((((((((((...(....)..))))..)))))))))))))))))))))) ( -30.90, z-score = -3.63, R) >droSec1.super_1 4494309 90 - 14215200 ------------CUCCCGCUUGCUGUUGUUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGU ------------.....((((((((((((((((((.........))).....(((((((((((...(....)..))))..)))))))))))))))))))))) ( -30.90, z-score = -3.63, R) >droYak2.chr2R 18922034 102 + 21139217 CUCCCGGUGGCUGUUGUUCUCGUUGUUGUUGCGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGU ..................(((((..((((((((((.........))))).(((((((((((((...(....)..))))..))))))))))))))..))))). ( -32.30, z-score = -2.25, R) >consensus ____________CUCCCGCUUGCUGUUGUUGUGACUGUAAAAGUGUCGUCAUUGUUGUUGCCACUGCUGCUGAAUGGCUUAACAACAAUAACAAUGGCGAGU .................((((((..((((((((((.........)))).)(((((((((((((...(....)..))))..))))))))))))))..)))))) (-30.64 = -30.32 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:11 2011