| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,918,352 – 6,918,442 |
| Length | 90 |
| Max. P | 0.906729 |

| Location | 6,918,352 – 6,918,442 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.61 |
| Shannon entropy | 0.57162 |
| G+C content | 0.43725 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
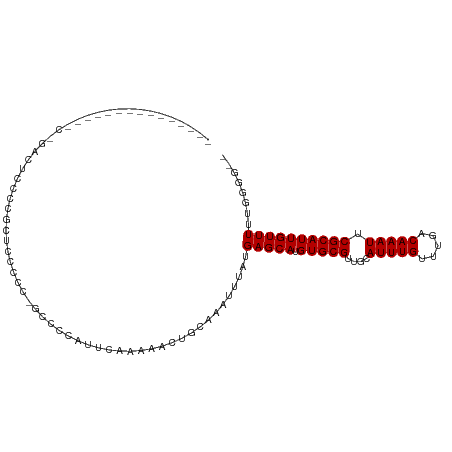
>dm3.chr2R 6918352 90 + 21146708 ---------------CUGAUUUUCCGCAUACCC--CCCCAUUCAAAAACUGCGAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUUUGCGU-- ---------------..................--.......((((((((((((((((..(((((.((((...)))).)))))...)))))))))..)))))))...-- ( -21.50, z-score = -2.31, R) >droSim1.chr2R 5459138 94 + 19596830 ---------------CCGAUUUCCCGCUUUUCUCGCCCCAUUCAAAAACUGCGAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUUUGGGGUU ---------------...................((((((....((((((((((((((..(((((.((((...)))).)))))...)))))))))..))))))))))). ( -27.80, z-score = -2.71, R) >droSec1.super_1 4485904 83 + 14215200 -----------------------CCGAUUUCCC-GCCCCAUUCAAAAACUGCAAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUUGGGGU-- -----------------------..........-((((((..(((....(((.(((((..(((((.((((...)))).)))))...))))).))))))...))))))-- ( -21.60, z-score = -1.16, R) >droYak2.chr2R 18913400 102 - 21139217 CCGAUUUUCCACUCUUCCGCUCCCUCCUCCAUUUUCUUAAUUCAAAAACUGCGAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUU------- ............................................((((((((((((((..(((((.((((...)))).)))))...)))))))))..)))))------- ( -17.80, z-score = -2.01, R) >droEre2.scaffold_4845 3712704 85 - 22589142 ---------------CCGAUUUUCCGCGUUUCC--CCCCAUUCAAAAACUGCGAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUU------- ---------------..................--.........((((((((((((((..(((((.((((...)))).)))))...)))))))))..)))))------- ( -17.80, z-score = -1.55, R) >droAna3.scaffold_13266 624757 93 + 19884421 ------------AUCCUGGCCCAGUUUUCCCUCUAACUCGCUGUAUAG--GAAAAUUUAUGAGCAUGUGCGUUGCAUUUGCUUGACAAAUUCGCAUUGUUUUUGCAU-- ------------.(((((...((((..............))))..)))--))........(((((.((((...)))).))))).........(((.......)))..-- ( -18.94, z-score = -0.15, R) >dp4.chr3 947378 90 - 19779522 -----------------CCCCCCCUUCGCCCCACGCCCCCUUCCACCAGGGGAAAUUUAUGAGCAUGUGCGUUUCAUUUGGUUGACAAAUUCGCAUUGUUUUUGGGG-- -----------------...........(((((...(((((......)))))........(((((.(((((....(((((.....))))).)))))))))).)))))-- ( -26.80, z-score = -1.23, R) >droPer1.super_34 132368 81 - 916997 --------------------------CCCCCCAUGCCCCCUUCCACCAGGGGAAAUUUAUGAGCAUGUGCGUUUCAUUUGGUUGACAAAUUCGCAUUGUUUUUGGGG-- --------------------------..(((((...(((((......)))))........(((((.(((((....(((((.....))))).)))))))))).)))))-- ( -27.00, z-score = -1.90, R) >consensus _______________C_GACUCCCCGCUCCCCC_GCCCCAUUCAAAAACUGCAAAUUUAUGAGCAUGUGCGUUGCAUUUGUUUGACAAAUUCGCAUUGUUUUUGGGG__ ............................................................(((((.(((((....(((((.....))))).))))))))))........ (-10.43 = -10.43 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:08 2011