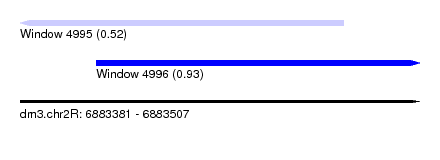
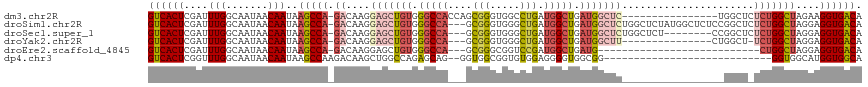
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,883,381 – 6,883,507 |
| Length | 126 |
| Max. P | 0.930996 |

| Location | 6,883,381 – 6,883,483 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Shannon entropy | 0.41015 |
| G+C content | 0.52207 |
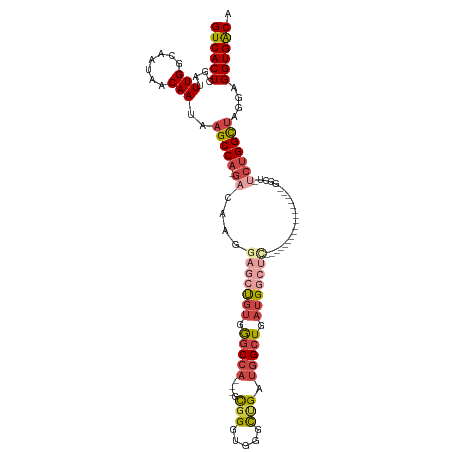
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -16.41 |
| Energy contribution | -17.61 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
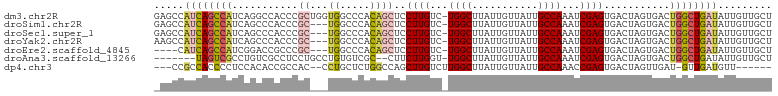
>dm3.chr2R 6883381 102 - 21146708 GAGCCAUCAGCCAUCAGGCCACCCGCUGGUGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU .....((((((((...(((((((....)))))))(((((.((((((..-((((...........))))...)))).)))).)))..))))))))......... ( -39.50, z-score = -2.85, R) >droSim1.chr2R 5438586 99 - 19596830 GAGCCAUCAGCCAUCAGCCCACCCGC---UGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU .....(((((((((((((......))---)))..(((((.((((((..-((((...........))))...)))).)))).)))..))))))))......... ( -30.60, z-score = -1.50, R) >droSec1.super_1 4465255 99 - 14215200 GAGCCAUCAGCCAUCAGCCCACCCGC---UGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU .....(((((((((((((......))---)))..(((((.((((((..-((((...........))))...)))).)))).)))..))))))))......... ( -30.60, z-score = -1.50, R) >droYak2.chr2R 18891691 99 + 21139217 AAGCCAUCAGCCAUCAGCCCACCCGC---UGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU .....(((((((((((((......))---)))..(((((.((((((..-((((...........))))...)))).)))).)))..))))))))......... ( -30.60, z-score = -1.70, R) >droEre2.scaffold_4845 3692703 95 + 22589142 ----CAUCAGCCAUCGGACCGCCCGC---UGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU ----.((((((((((((.....))((---((.....))))((((((..-((((...........))))...)))).)).....)).))))))))......... ( -29.20, z-score = -1.27, R) >droAna3.scaffold_13266 602658 93 - 19884421 -------UAGUCGCCUGUCGCCUCCUGCCUGUGUCGC--CUUCUUGGU-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU -------..((((((.(((((.....))(((.(((((--....(((((-((((...........))).)))))))))))))).))).))).)))......... ( -24.20, z-score = -1.25, R) >dp4.chr3 925602 91 + 19779522 ---CCGCCACCCCUCCACACCGCCAC--CCUGCUCUGGCCAGCUUGUCUUGGCUUAUUGUUAUUGCCAAACCGAGUGACUAGUUGAU-GUUGAUGUU------ ---..........((.(((.(((((.--.......))))..(((((..(((((...........)))))..)))))........).)-)).))....------ ( -18.50, z-score = -0.17, R) >consensus _AGCCAUCAGCCAUCAGCCCACCCGC___UGGCCCACAGCUCCUUGUC_UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUAUUGUUGCU .....((((((((.........((......))..(((...((((((...((((...........))))...)))).))...)))..))))))))......... (-16.41 = -17.61 + 1.21)
| Location | 6,883,405 – 6,883,507 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Shannon entropy | 0.40778 |
| G+C content | 0.57378 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -23.68 |
| Energy contribution | -25.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
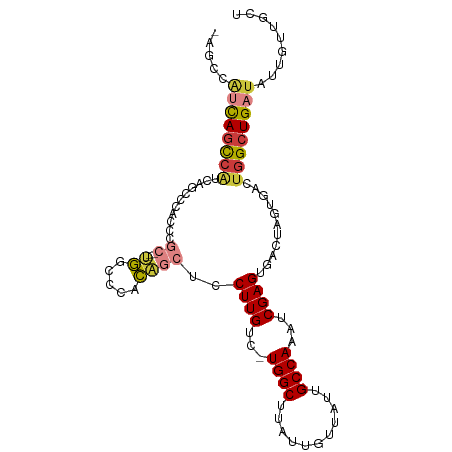
>dm3.chr2R 6883405 102 + 21146708 GUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCACCAGCGGGUGGCCUGAUGGCUGAUGGCUC----------------UGGCUCUCUGGCUAGAAGGUGACA ((((((...((((((..........(((((-(((....((((((((((((((....)))))))).))))))....).))----------------))))).....)))))).)))))). ( -45.26, z-score = -3.11, R) >droSim1.chr2R 5438610 115 + 19596830 GUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCA---GCGGGUGGGCUGAUGGCUGAUGGCUCUGGCUCUAUGGCUCUCCGGCUCUCUGGCUAGGAGGUGACA (((((....................(((((-.....((((((((.(((((---.(((.....))).))))).)))))))).......)))))((((((((....)))).))))))))). ( -46.60, z-score = -1.44, R) >droSec1.super_1 4465279 107 + 14215200 GUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCA---GCGGGUGGGCUGAUGGCUGAUGGCUCUGGCUCU--------CCGGCUCUCUGGCUAGGAGGUGACA (((((..(.((((((...........))))-)))..((((((((.(((((---.(((.....))).))))).))))))))....((--------((((((....)))).))))))))). ( -44.80, z-score = -1.83, R) >droYak2.chr2R 18891715 99 - 21139217 GUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCA---GCGGGUGGGCUGAUGGCUGAUGGCUU---------------CUGGCU-UCUGGCUAGGAGGUGACA (((((((...(((.......)))..(((((-((..(((((((((.(((((---.(((.....))).))))).)))))))---------------))....-)))))))..)).))))). ( -43.90, z-score = -3.65, R) >droEre2.scaffold_4845 3692727 88 - 22589142 GUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCA---GCGGGCGGUCCGAUGGCUGAUG---------------------------CUGGCUAGGAGGUGACA ((((((.(.((((((...........))))-)))......((...(((((---(((..(((((....))))).))---------------------------)))))).)).)))))). ( -34.80, z-score = -2.33, R) >dp4.chr3 925619 89 - 19779522 GUCACUCGGUUUGGCAAUAACAAUAAGCCAAGACAAGCUGGCCAGAGCAG--GGUGGCGGUGUGGAGGGGUGGCGG----------------------------GGUGGCAUGGUGGCA (((((((.(((((((...........))))).(((.((((.((......)--).))))..))).........)).)----------------------------))))))......... ( -27.30, z-score = 0.01, R) >consensus GUCACUCGAUUUGGCAAUAACAAUAAGCCA_GACAAGGAGCUGUGGGCCA___GCGGGUGGGCUGAUGGCUGAUGGCUC_________________GGCU_UCUGGCUAGGAGGUGACA ((((((.(...((((...........))))...)...(((((((.(((((....(((.....))).))))).))))))).................................)))))). (-23.68 = -25.13 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:14:02 2011