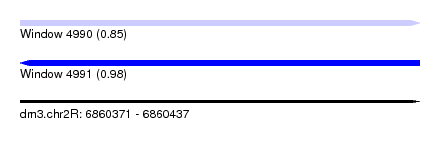
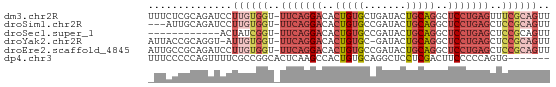
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,860,371 – 6,860,437 |
| Length | 66 |
| Max. P | 0.975929 |

| Location | 6,860,371 – 6,860,437 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.61727 |
| G+C content | 0.55063 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -9.04 |
| Energy contribution | -10.07 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
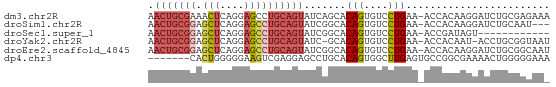
>dm3.chr2R 6860371 66 + 21146708 AACUGCGAAACUCAGGAGCCUGCAGUAUCAGCACAGUGUCCUGAA-ACCACAAGGAUCUGCGAGAAA ...(((((.((((((....))).))).)).)))(((.(((((...-......))))))))....... ( -16.30, z-score = -0.58, R) >droSim1.chr2R 5417457 63 + 19596830 AACUGCGGAGCUCAGGAGCCUGCAGUAUCGGCACAGUGUCCUGAA-ACCACAAGGAUCUGCAAU--- .(((((((.(((....)))))))))).......(((.(((((...-......))))))))....--- ( -21.40, z-score = -1.62, R) >droSec1.super_1 4442993 54 + 14215200 AACUGCGGAGCUCAGGAGCCUGCAGUAUCGGCACAGUGUCCUGAA-ACCGAUAGU------------ .(((((((.(((....))))))))))(((((..(((....)))..-.)))))...------------ ( -21.30, z-score = -2.69, R) >droYak2.chr2R 18868381 64 - 21139217 AACUGCGGAGCUCAGGAGCCUGCAGUAUC-GCACAGUGUCCUGAA-ACCACAAU-ACCUGCGGUAAU .(((((((.(((....))))))))))(((-(((..((((..((..-..))..))-)).))))))... ( -23.20, z-score = -2.72, R) >droEre2.scaffold_4845 3667212 66 - 22589142 AACUGCGGAGCUCAGGAGCCUGCAGUAUCGGCACAGUGUCCUGAA-ACCACAAGGAUCUGCGGCAAU .(((((((.(((....))))))))))...(.(.(((.(((((...-......)))))))).).)... ( -25.00, z-score = -2.07, R) >dp4.chr3 902910 60 - 19779522 -------CACUGGGGGAAGUCGAGGAGCCUGCACAGUGGCUUGAGUGCCGGCGAAAACUGGGGGAAA -------..(..(.....((((.((((((........))))).....))))).....)..)...... ( -13.60, z-score = 1.40, R) >consensus AACUGCGGAGCUCAGGAGCCUGCAGUAUCGGCACAGUGUCCUGAA_ACCACAAGGAUCUGCGGGAA_ .(((((((.(((....)))))))))).......(((....)))........................ ( -9.04 = -10.07 + 1.03)
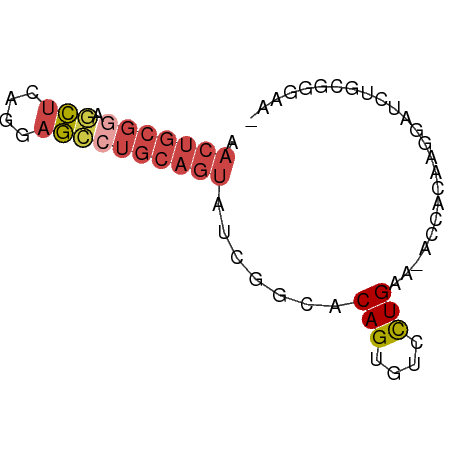
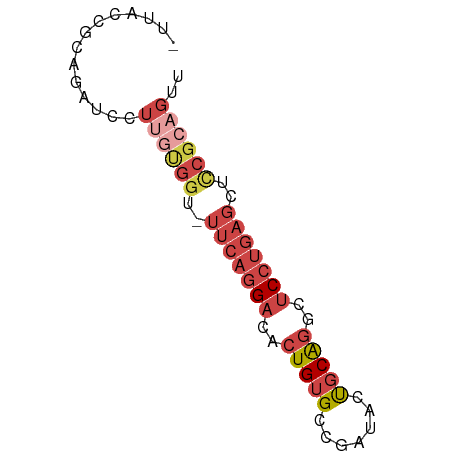
| Location | 6,860,371 – 6,860,437 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.61727 |
| G+C content | 0.55063 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -11.91 |
| Energy contribution | -13.92 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6860371 66 - 21146708 UUUCUCGCAGAUCCUUGUGGU-UUCAGGACACUGUGCUGAUACUGCAGGCUCCUGAGUUUCGCAGUU ..............((((((.-(((((((..((((.........))))..)))))))..)))))).. ( -18.70, z-score = -0.46, R) >droSim1.chr2R 5417457 63 - 19596830 ---AUUGCAGAUCCUUGUGGU-UUCAGGACACUGUGCCGAUACUGCAGGCUCCUGAGCUCCGCAGUU ---...........((((((.-(((((((..((((.........))))..)))))))..)))))).. ( -21.10, z-score = -1.21, R) >droSec1.super_1 4442993 54 - 14215200 ------------ACUAUCGGU-UUCAGGACACUGUGCCGAUACUGCAGGCUCCUGAGCUCCGCAGUU ------------..(((((((-..(((....))).)))))))((((.((((....))))..)))).. ( -21.50, z-score = -2.83, R) >droYak2.chr2R 18868381 64 + 21139217 AUUACCGCAGGU-AUUGUGGU-UUCAGGACACUGUGC-GAUACUGCAGGCUCCUGAGCUCCGCAGUU ..((((...)))-)((((((.-(((((((..((((..-......))))..)))))))..)))))).. ( -23.50, z-score = -2.01, R) >droEre2.scaffold_4845 3667212 66 + 22589142 AUUGCCGCAGAUCCUUGUGGU-UUCAGGACACUGUGCCGAUACUGCAGGCUCCUGAGCUCCGCAGUU ((((.(((((.((((.(....-..)))))..))))).))))(((((.((((....))))..))))). ( -21.30, z-score = -0.75, R) >dp4.chr3 902910 60 + 19779522 UUUCCCCCAGUUUUCGCCGGCACUCAAGCCACUGUGCAGGCUCCUCGACUUCCCCCAGUG------- ...............(((.((((..........)))).)))...................------- ( -11.00, z-score = -0.32, R) >consensus _UUACCGCAGAUCCUUGUGGU_UUCAGGACACUGUGCCGAUACUGCAGGCUCCUGAGCUCCGCAGUU ..............((((((...((((((..(((((.......)))))..))))))...)))))).. (-11.91 = -13.92 + 2.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:58 2011