
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,515,497 – 2,515,599 |
| Length | 102 |
| Max. P | 0.948192 |

| Location | 2,515,497 – 2,515,599 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.09 |
| Shannon entropy | 0.63843 |
| G+C content | 0.35627 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -10.26 |
| Energy contribution | -9.90 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2515497 102 + 23011544 CUACAUACUCAUUAGGCACUCUGAAAUGCGCCUAGUCAGGGAAAAUGUAU-AUUCAAUUUGCACAGUUUCGUUAAUAUACAUGUGGGUAAAAUAGAGAGAGGA-------- ((((((.((.(((((((.(........).))))))).)).((((.((((.-........))))...))))..........)))))).................-------- ( -20.00, z-score = -0.21, R) >droSim1.chr2L 2478120 105 + 22036055 CUACAUACUCAUUAGGCACUCUGAAAUGCGCCUAACCAGGGAAAAUGUA---UUCAAUUUGGACGUUUUCGUUAAAAUACGUGUAUUUUAGACACACGGAAAAUAUGA--- ...((((....((((((.(........).))))))((...((((((((.---..........))))))))(((((((((....)))))).)))....))....)))).--- ( -21.70, z-score = -1.40, R) >droSec1.super_5 689643 105 + 5866729 CUACAUACUCAUUAGGCACUCUGAAAUGCGCCUAACCAGGGAAAAUGUA---UUCAAUUUGUACGUUUUCGUUAAAAUACGUGUAUUUUAGACACACGGAAAAUAGGA--- ...........((((((.(........).))))))((((.(((((((((---(.......)))))))))).))......(((((.........))))).......)).--- ( -24.70, z-score = -2.33, R) >droEre2.scaffold_4929 2561616 96 + 26641161 CUACAUACUCAUUAGGCACUCUGAAAUGCGCCUAGCCAGGGAAAAUGUA---UUCAAUUUGCACGUUUUCGUUGAAAUAUGUAUGUCUAAAACACAAAA------------ ..((((((...((((((.(........).)))))).(((.((((((((.---..........)))))))).)))......)))))).............------------ ( -22.00, z-score = -2.17, R) >droVir3.scaffold_12963 9899778 103 + 20206255 --------AACUUGUGUUCCCAAAAUUGAUUGCACCUUUGGAAUAUGUUUGAGUUAAAAUGAACUUCUUUAGUUGACAACUUUUAUAGUGAGCAAUCAGGAAAAGAGAACU --------..(((...((((......(((((((.(((.(((((..((((.(((((......)))))........))))..))))).)).).)))))))))))..))).... ( -18.70, z-score = -0.31, R) >consensus CUACAUACUCAUUAGGCACUCUGAAAUGCGCCUAACCAGGGAAAAUGUA___UUCAAUUUGCACGUUUUCGUUAAAAUACGUGUAUUUUAAACACACAGAAAA___GA___ .......((..((((((.(........).))))))..)).((((((((..............))))))))......................................... (-10.26 = -9.90 + -0.36)

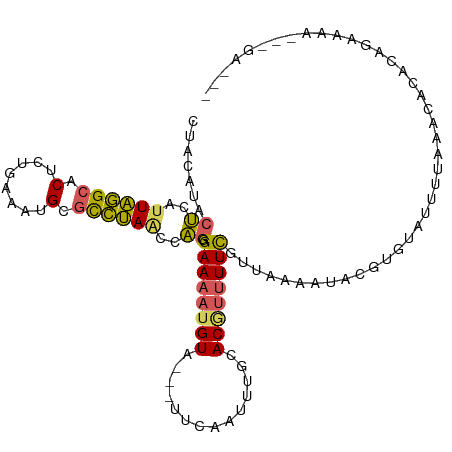
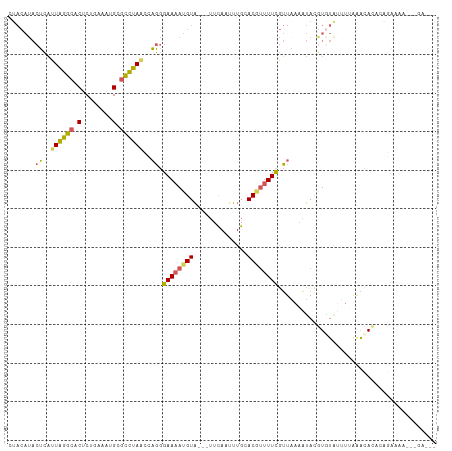
| Location | 2,515,497 – 2,515,599 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 64.09 |
| Shannon entropy | 0.63843 |
| G+C content | 0.35627 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -8.90 |
| Energy contribution | -10.90 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2515497 102 - 23011544 --------UCCUCUCUCUAUUUUACCCACAUGUAUAUUAACGAAACUGUGCAAAUUGAAU-AUACAUUUUCCCUGACUAGGCGCAUUUCAGAGUGCCUAAUGAGUAUGUAG --------........((((..(((.((.(((((((((.(((....)))........)))-))))))..........(((((((........))))))).)).))).)))) ( -18.40, z-score = -1.24, R) >droSim1.chr2L 2478120 105 - 22036055 ---UCAUAUUUUCCGUGUGUCUAAAAUACACGUAUUUUAACGAAAACGUCCAAAUUGAA---UACAUUUUCCCUGGUUAGGCGCAUUUCAGAGUGCCUAAUGAGUAUGUAG ---(((.......(((((((.....))))))).((((..(((....)))..))))))).---(((((.....((.(((((((((........))))))))).)).))))). ( -25.80, z-score = -3.08, R) >droSec1.super_5 689643 105 - 5866729 ---UCCUAUUUUCCGUGUGUCUAAAAUACACGUAUUUUAACGAAAACGUACAAAUUGAA---UACAUUUUCCCUGGUUAGGCGCAUUUCAGAGUGCCUAAUGAGUAUGUAG ---..........(((((((.....))))))).............((((((.....(((---......)))..(.(((((((((........))))))))).))))))).. ( -25.90, z-score = -2.98, R) >droEre2.scaffold_4929 2561616 96 - 26641161 ------------UUUUGUGUUUUAGACAUACAUAUUUCAACGAAAACGUGCAAAUUGAA---UACAUUUUCCCUGGCUAGGCGCAUUUCAGAGUGCCUAAUGAGUAUGUAG ------------.............((((((.....(((..(((((.(((.........---))).)))))..))).(((((((........)))))))....)))))).. ( -19.70, z-score = -0.81, R) >droVir3.scaffold_12963 9899778 103 - 20206255 AGUUCUCUUUUCCUGAUUGCUCACUAUAAAAGUUGUCAACUAAAGAAGUUCAUUUUAACUCAAACAUAUUCCAAAGGUGCAAUCAAUUUUGGGAACACAAGUU-------- .((((((......(((((((.(.((.....(((.....)))...((.(((......))))).............))).))))))).....)))))).......-------- ( -17.50, z-score = -0.92, R) >consensus ___UC___UUUUCUGUGUGUUUAAAAUACACGUAUUUUAACGAAAACGUGCAAAUUGAA___UACAUUUUCCCUGGCUAGGCGCAUUUCAGAGUGCCUAAUGAGUAUGUAG .............................((((((((....(((((.(((............))).)))))......(((((((........)))))))..)))))))).. ( -8.90 = -10.90 + 2.00)

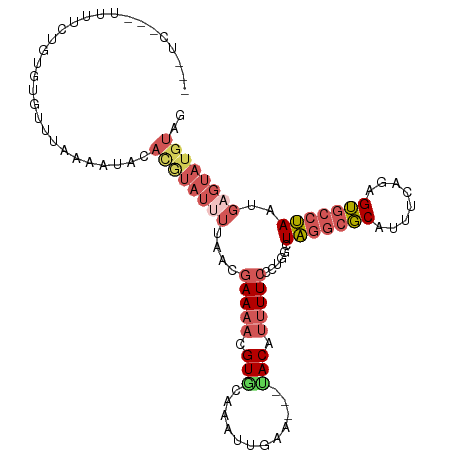
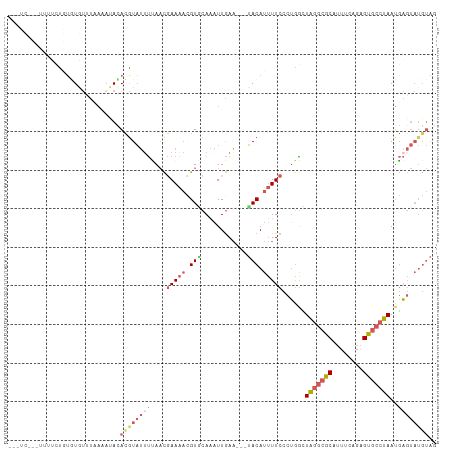
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:34 2011