
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,832,166 – 6,832,300 |
| Length | 134 |
| Max. P | 0.660281 |

| Location | 6,832,166 – 6,832,300 |
|---|---|
| Length | 134 |
| Sequences | 12 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Shannon entropy | 0.50975 |
| G+C content | 0.40963 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6832166 134 + 21146708 UGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCAAGCGCAGACAAACGCCCAGAGAAAACAAGUGAAUGACG-CUGACUU-GGCGCAAUCGAAGUUCGGUUACUCUUCA------ ....(((((.-...(((((((..((((....(((.....))).(((((......-.)))))))))..)))))))..(((..(((......((((.....))-))..)))-)))))(((((.....))))).)))....------ ( -34.80, z-score = -0.44, R) >droSim1.chr2R 5400835 134 + 19596830 UGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAAACGAUAAAA-ACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAAGUGAAUGACG-CUGACUG-GGCGCAAUCGAAGUUCGGUUACUCUUCA------ ....(((((.-...(((((((..(((((((.(((.....))).))))(((....-..)))..)))..)))))))..((((((........((((.....))-))..)))-)))))(((((.....))))).)))....------ ( -37.10, z-score = -1.28, R) >droSec1.super_1 4422504 134 + 14215200 UGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAAGUGAAUGACG-CUGACUU-GGCGCAAUCGAAGUUCGGUUACUCUUCA------ ...((.((((-...(((((((..((((....(((.....))).(((((......-.)))))))))..))))))).))))))(((.............(.((-((.....-)))))(((((.....))))).)))....------ ( -36.40, z-score = -1.01, R) >droYak2.chr2R 18847275 134 - 21139217 UGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCAAGCGCAGACAAAUGCCCAGAUAAAACAAGUGAAUGACG-CUGACUU-GGCGCAAUCGAAGUUCGGUUACUCUUCA------ ((((..((((-...(((((((..((((....(((.....))).(((((......-.)))))))))..))))))).))))..))))........((.((.((-((.....-)))))).))((((..........)))).------ ( -36.50, z-score = -1.23, R) >droEre2.scaffold_4845 3646694 134 - 22589142 UGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAAGUGAAUGACG-CUGACUU-GGCGCAAUCGAAGUUCGGUUACUCUUCA------ ...((.((((-...(((((((..((((....(((.....))).(((((......-.)))))))))..))))))).))))))(((.............(.((-((.....-)))))(((((.....))))).)))....------ ( -36.40, z-score = -1.01, R) >droAna3.scaffold_13266 562221 134 + 19884421 UGUUGAGACA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAACUGACUGA-GCCUGACUU-GGCACAAUCGAACUUUGGUUGCUCUUCG------ ....(((.((-(..(((((((..((((....(((.....))).(((((......-.)))))))))..)))))))....((((((....((......)).-(((......-))).........))))))))))))....------ ( -35.30, z-score = -1.42, R) >dp4.chr3 886877 137 - 19779522 UGUUGGGACAGAAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAGGCGGAUGACGGCCGACUUUAGAGCCAUCGAAGAGGAGAUGCUCUUUG------ (((((((...((...(((((....((((((.(((.....))).))))))...))-)))..))...).)))))).((((((....((((..(((.(....).)))..)))).).)))))(((((((......)))))))------ ( -43.10, z-score = -1.81, R) >droPer1.super_34 73099 137 - 916997 UGUUGGGACAGAAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-ACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAGGCGGAUGACGGCCGACUUUAGAGCCAUCGAAGAGGGGAUGCUCUUUG------ (((((((...((...(((((....((((((.(((.....))).))))))...))-)))..))...).)))))).((((((....((((..(((.(....).)))..)))).).)))))(((((((......)))))))------ ( -43.10, z-score = -1.53, R) >droWil1.scaffold_180697 3153625 135 + 4168966 UAUCGAGACAAAAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACAAUAAAAACCAUUUCAAGUGCAGACAG-CAAAAAUAAUGCUUCAA--AUCAAUUUGCCAGCGGCAAGAUAACGAAAUUGCUAUACUCUUGG------ ...(((((...((((.((((....((((((.(((.....))).))))))...)))).)))).....(((((.((-((.......))))))..--.....((((((...))))))..........))).....))))).------ ( -27.60, z-score = -0.81, R) >droVir3.scaffold_12823 2147842 131 - 2474545 UAUCGAGAUAAAA-UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-CCGUCUCAAGCGUAGACAAAUGCCAAGAGUAAACCAAG-A--AAAUUUGUAAAUAUAAUAUAAUGAAUUUGGCAUACUCUC-------- .............-......(((((((((((.((((.......(((((......-.))))).)))).)))))))))))..((((((..(((((-.--....(((((.......)))))....)))))..)))))).-------- ( -30.50, z-score = -3.08, R) >droMoj3.scaffold_6496 26696289 141 - 26866924 GCUCGAGACAAAA-UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA-CCGUCUCAAGCGCAGACAAAUGCCAACAAUAAGCCAAG-AACAAAUUUAUGCACUUAAUACACUGAAUUUGAUAUACCCUUACUCCCUA ....(((.....(-(((((.((((((((((.(((.....))).(((((......-.))))).......)))))))))).))))))........-..((((((((((.........)).))))))))...........))).... ( -31.00, z-score = -2.98, R) >droGri2.scaffold_15245 6315910 132 + 18325388 CAUCGAAAGAAAA-UGGUUUGUAUUUGCUUAGGCUUAUGGCCAAGAAGAUAAAA-CCGUCUCAAGCACAGACAAAUGCCAACAAUAAACCAAC-AAUAAAGUUUUGCUUAUAAUAAAGGGAAUUUGAUAUACUCU--------- .((((((......-((((((((...(((((.(((.....)))..((.(((....-..))))))))))...))))..)))).............-........(((.(((......))).))))))))).......--------- ( -23.30, z-score = -0.52, R) >consensus UGUUGAGACA_AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAA_ACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAAGUGAAUGACG_CUGACUU_GGAGCAAUCGAAGUUGGGAUACUCUUCA______ ..............(((((((..((((....(((.....))).(((((........)))))))))..)))))))...................................................................... (-16.83 = -17.06 + 0.22)

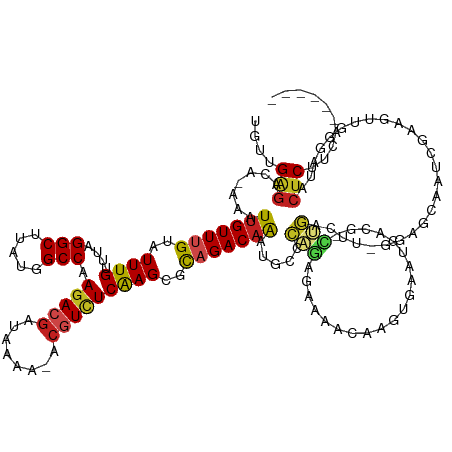

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:49 2011