| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,510,785 – 2,510,878 |
| Length | 93 |
| Max. P | 0.697529 |

| Location | 2,510,785 – 2,510,878 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Shannon entropy | 0.34204 |
| G+C content | 0.37370 |
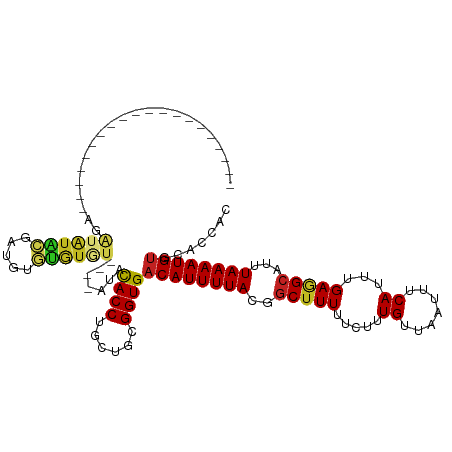
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
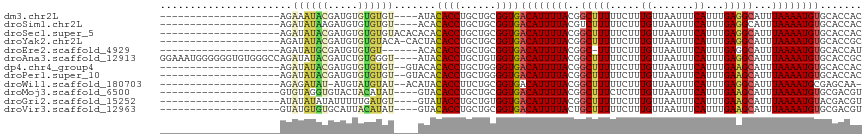
>dm3.chr2L 2510785 93 - 23011544 --------------------AGAAAUACGAUGUGUGUGU----AUACACCUGCUGCGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCAC --------------------...........(((.((((----...((((......))))((((((((..(((((.....((.......))...)))))...))))))))))))))) ( -21.50, z-score = -1.04, R) >droSim1.chr2L 2473476 93 - 22036055 --------------------AGAUAUAAGAUGUGUGUGU----ACACACCUGCUGCGGUGACAUUUUACGUCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCAC --------------------.......((..(((((...----.)))))...))..((((((((((((.(((((......((.......))...)))))...)))))))).)))).. ( -21.70, z-score = -1.25, R) >droSec1.super_5 685014 97 - 5866729 --------------------AGAUAUACGAUGUGUGUGUACACACACACCUGCUGCGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCAC --------------------...........(((((((....))))))).......((((((((((((..(((((.....((.......))...)))))...)))))))).)))).. ( -27.50, z-score = -2.62, R) >droYak2.chr2L 2503215 96 - 22324452 --------------------AGAUAUACGAUGUGUGUACA-CACUACACCUGCUGCGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCGC --------------------........(.((((((....-))).))).)....((((((((((((((..(((((.....((.......))...)))))...)))))))).)))))) ( -24.90, z-score = -2.12, R) >droEre2.scaffold_4929 2556863 90 - 26641161 --------------------AGAUAUGCGAUGUGUGU------ACACACCUGCUGCGGUGACAUUUUACGGC-UUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCAU --------------------......(((.((((...------.))))..)))...((((((((((((..((-(((....((.......))...)))))...)))))))).)))).. ( -23.60, z-score = -1.59, R) >droAna3.scaffold_12913 264869 113 + 441482 GGAAAUGGGGGGUGUGGGCCAGAUAUACGAUCUGUGGGU----AUACACCUGCUGUGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCGC ......((.(((((((.(((((((.....))))...)))----.))))))).))((((((((((((((..(((((.....((.......))...)))))...)))))))).)))))) ( -36.80, z-score = -2.75, R) >dp4.chr4_group4 1794467 95 - 6586962 --------------------AGAUAUACGAUGUGUGUGU--GUACACACCUGCUGGGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAAGCAUUUAAAAUGUGCACCAC --------------------...........((((((..--..)))))).......((((((((((((..(((((.....((.......))...)))))...)))))))).)))).. ( -23.70, z-score = -1.33, R) >droPer1.super_10 796663 95 - 3432795 --------------------AGAUAUACGAUGUGUGUGU--GUACACACCUGCUGGGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAAGCAUUUAAAAUGUGCACCAC --------------------...........((((((..--..)))))).......((((((((((((..(((((.....((.......))...)))))...)))))))).)))).. ( -23.70, z-score = -1.33, R) >droWil1.scaffold_180703 881585 93 + 3946847 --------------------AGAGAUAU-AUGUAUGUAU--ACAUACACCUUCUGCGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGCGAGCAA- --------------------..(((...-.((((((...--.))))))...)))((..((.(((((((..(((((.....((.......))...)))))...))))))))).))..- ( -18.80, z-score = -1.17, R) >droMoj3.scaffold_6500 31002386 93 + 32352404 --------------------GUGUAGGUGUACUACAUAU----GUACACCUGCUGCGGUGACAUUUUACGGCUUUCUCUUUGUUAAUUUCAUUUGAAGCAUUUAAAAUGUGCGACGU --------------------..((((((((((.......----)))))))))).(((.((((((((((..(((((.....((.......))...)))))...)))))))).)).))) ( -28.60, z-score = -3.25, R) >droGri2.scaffold_15252 5279159 93 + 17193109 --------------------AUAUAUAUAUUUUUGAUGU----GUAUACCUGCUGUGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAAGCAUUUAAAAUGUACGACGU --------------------.((((((((((...)))))----)))))......((.(((.(((((((..(((((.....((.......))...)))))...)))))))))).)).. ( -19.20, z-score = -1.95, R) >droVir3.scaffold_12963 9892467 93 - 20206255 --------------------GUAUGUGUGCAUUACAUAU----GUACACCUGCUGCGGUGACAUUUUACUGCUUUUUCUUUGUUAAUUUCAUUUGAAGCAUUUAAAAUGUGCGACGU --------------------(((.((((((((.....))----)))))).))).(((.((((((((((.((((((.....((.......))...))))))..)))))))).)).))) ( -25.40, z-score = -2.59, R) >consensus ____________________AGAUAUACGAUGUGUGUGU____AUACACCUGCUGCGGUGACAUUUUACGGCUUUUUCUUUGUUAAUUUCAUUUGAGGCAUUUAAAAUGUGCACCAC ......................((((((.....)))))).......((((......))))((((((((..(((((.....((.......))...)))))...))))))))....... (-14.55 = -14.38 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:33 2011