| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,778,466 – 6,778,583 |
| Length | 117 |
| Max. P | 0.633352 |

| Location | 6,778,466 – 6,778,583 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.52 |
| Shannon entropy | 0.61685 |
| G+C content | 0.49252 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -11.10 |
| Energy contribution | -10.64 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
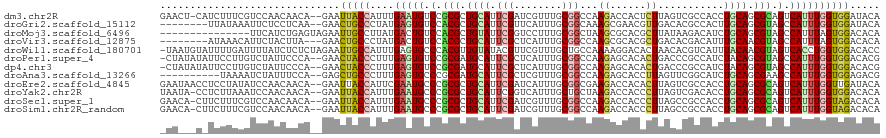
>dm3.chr2R 6778466 117 + 21146708 GAACU-CAUCUUUCGUCCAACAACA--GAAUUACCAUUUGAAUGUCCGCGCUGCAUUCGAUCGUUUGCGGCCAAGACCACUCUUAGUCGCCACCUGCAGCGCAGUCAUUUGGUGGAUACA .....-...................--......(((.(..((((.(.((((((((...((....))(((((.((((....)))).)))))....)))))))).).))))..))))..... ( -33.20, z-score = -2.02, R) >droGri2.scaffold_15112 4199580 110 - 5172618 --------UUAUAAAUUCUCCUCAA--GAACUGCCCUAUGAGUGUCCACGCUGCAUUCGUUCAUUUGCGGCAAAGCGAACGUUGACACGCCACUUGCAGCGUAGCCAUUUGGUGGAUACA --------.......((((.....)--)))...........((((((((((((((..(((((((((((......)))))...))).))).....)))))))).(((....))))))))). ( -32.40, z-score = -1.65, R) >droMoj3.scaffold_6496 23970699 105 + 26866924 ---------------UUCAUCUGAGUAGAAUUGCCUUAUGACUGUCCACGCUGUAUUCGGUCCUUUGCGGCUAAGCGCACGCUUAUAAGACAUCUGCAGCGUAGCCAUUUAGUGGACACA ---------------.((((..(.(((....))))..)))).(((((((((((((..........))))))...(.(((((((....((....))..))))).))).....))))))).. ( -28.80, z-score = -0.54, R) >droVir3.scaffold_12875 14540417 109 - 20611582 --------AUAAACAUUCUACUUA---GAACUGCCCUAUGACUGUCCACGCUGCAUUCGCUCAUUUGCGGCCAAGCGCACGCUGACACGACAUUUGCAACGUAGCCAUUUAGUGGACACA --------.......((((....)---)))............(((((((((((((..........))))))...(.(((((.((.((.......)))).))).))).....))))))).. ( -25.10, z-score = -0.54, R) >droWil1.scaffold_180701 2705435 119 + 3904529 -UAAUGUAUUUUGAUUUUAUCUCUCUAGAAUUGCCAUUUGAGUGCCCACGUUGUAUACGUUCGUUUGCUGCCAAAAGGACACUAACACGUCAUUUACAACGUAGUCACCUGGUGGACACC -...(((.....(((((((......))))))).((((..(.(((.(.((((((((.((((..(((.(.((((....)).))).)))))))....)))))))).).))))..))))))).. ( -26.70, z-score = -0.84, R) >droPer1.super_4 1859468 117 + 7162766 -CUAUAUAUUCCUUGUCUAUUCCCA--GAACUACCCUUUGAGUGUCCGCGAUGCAUUCGCUCAUUUGCGGCCAAGAGCACACUGACCCGCCAUCUACAGCGUAGCCAUUUGGUGGACACG -........................--..............((((((((((((....((((.....((((...((......))...)))).......))))....))))..)))))))). ( -26.40, z-score = -0.37, R) >dp4.chr3 7948364 117 + 19779522 -CUAUAUAUUCCUUGUCUAUUCCCA--GAACUACCCUUUGAGUGUCCGCGAUGCAUUCGCUCAUUUGCGGCCAAGAGCACACUGACCCGCCAUCUACAGCGUAGCCAUUUGGUGGACACG -........................--..............((((((((((((....((((.....((((...((......))...)))).......))))....))))..)))))))). ( -26.40, z-score = -0.37, R) >droAna3.scaffold_13266 6882598 108 - 19884421 ----------UAAAAUCUAUUUCCA--GAGCUGCCCUUUGAGUGCCCGCGAUGCAUUCGCUCGUUUGCGGCCAAGAGCACCUUAGUUCGGCAUCUGCAGCGAAGCCAUUUGGUGGAGACG ----------.........(((((.--..(((((.....((.(((((((((.((....))....))))).....((((......)))))))))).)))))...(((....)))))))).. ( -32.40, z-score = -0.19, R) >droEre2.scaffold_4845 3592524 118 - 22589142 GAAUAACCUCCUAUAUCCAACAACA--GAAUUACCAUUCGAAUGCCCGCGCUGCAUUCGAUCAUUUGCGGCGAAGACCACACUUAGUCGCCACCUGCAGCGCAGUCAUUUGGUUGAUACA ...(((((.................--((((....))))(((((.(.((((((((..(((....))).(((((..(.......)..)))))...)))))))).).))))))))))..... ( -31.00, z-score = -2.70, R) >droYak2.chr2R 18788710 117 - 21139217 UAAUA-CCUCUUAAAUCCAACAACA--GAAUUACCAUUUGAAUGCCCGCGCUGCAUUCGGUCAUUUGCUGCUAAGACCACCCUUAGUCGACACCUGCAGCGCAGUCAUUUGGUGGACACA .....-...................--......(((.(..((((.(.((((((((...(((...(((..((((((......))))))))).))))))))))).).))))..))))..... ( -31.80, z-score = -2.81, R) >droSec1.super_1 4365470 117 + 14215200 GAACA-CUUCUUUCGUCCAACAACA--GAAUUACCAUUUGAAUGCCCGCGCUGCAUUCGAUCGUUUGCGGCCAAGACCACCCUUAGCCGCCACCUGCAGCGCAGUCAUUUGGUAGACACA .....-...................--...((((((..(((......((((((((...((....))(((((.(((......))).)))))....))))))))..)))..))))))..... ( -33.20, z-score = -3.03, R) >droSim1.chr2R_random 1930572 117 + 2996586 GAACA-CUUCUUUCGUCCAACAACA--GAAUUACCAUUUGAAUGCCCGCGCUGCAUUCGAUCGUUUGCGGCCAAGACCACCCUUAGCCGCCACCUGCAGCGCAGUCAUUUGGUAGACACA .....-...................--...((((((..(((......((((((((...((....))(((((.(((......))).)))))....))))))))..)))..))))))..... ( -33.20, z-score = -3.03, R) >consensus _AA___C_UCUUAAAUCCAACAACA__GAAUUACCAUUUGAAUGCCCGCGCUGCAUUCGAUCAUUUGCGGCCAAGAGCACACUUACACGCCACCUGCAGCGUAGCCAUUUGGUGGACACA ..............................(((((....(((((.(.(((.((((.(((........)))...((......))...........)))).))).).))))))))))..... (-11.10 = -10.64 + -0.46)

| Location | 6,778,466 – 6,778,583 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.52 |
| Shannon entropy | 0.61685 |
| G+C content | 0.49252 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -16.44 |
| Energy contribution | -15.99 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
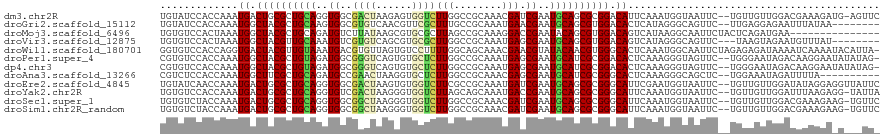
>dm3.chr2R 6778466 117 - 21146708 UGUAUCCACCAAAUGACUGCGCUGCAGGUGGCGACUAAGAGUGGUCUUGGCCGCAAACGAUCGAAUGCAGCGCGGACAUUCAAAUGGUAAUUC--UGUUGUUGGACGAAAGAUG-AGUUC ....(((((((((((.((((((((((.((((((((((....)))))...)))))...........)))))))))).))))....))))(((..--....))))))(....)...-..... ( -37.20, z-score = -1.33, R) >droGri2.scaffold_15112 4199580 110 + 5172618 UGUAUCCACCAAAUGGCUACGCUGCAAGUGGCGUGUCAACGUUCGCUUUGCCGCAAAUGAACGAAUGCAGCGUGGACACUCAUAGGGCAGUUC--UUGAGGAGAAUUUAUAA-------- ....(((.((....))((((((((((..(((....))).((((((.((((...))))))))))..)))))))))).........))).(((((--((...))))))).....-------- ( -30.00, z-score = 0.21, R) >droMoj3.scaffold_6496 23970699 105 - 26866924 UGUGUCCACUAAAUGGCUACGCUGCAGAUGUCUUAUAAGCGUGCGCUUAGCCGCAAAGGACCGAAUACAGCGUGGACAGUCAUAAGGCAAUUCUACUCAGAUGAA--------------- ..(((((((.....(....)((((.....(((((....(((.((.....)))))..)))))......)))))))))))(((....))).................--------------- ( -27.40, z-score = 0.06, R) >droVir3.scaffold_12875 14540417 109 + 20611582 UGUGUCCACUAAAUGGCUACGUUGCAAAUGUCGUGUCAGCGUGCGCUUGGCCGCAAAUGAGCGAAUGCAGCGUGGACAGUCAUAGGGCAGUUC---UAAGUAGAAUGUUUAU-------- ..(((((......((.((((((((((....((((.(((...((((......))))..))))))).)))))))))).))......)))))((((---(....)))))......-------- ( -36.60, z-score = -1.31, R) >droWil1.scaffold_180701 2705435 119 - 3904529 GGUGUCCACCAGGUGACUACGUUGUAAAUGACGUGUUAGUGUCCUUUUGGCAGCAAACGAACGUAUACAACGUGGGCACUCAAAUGGCAAUUCUAGAGAGAUAAAAUCAAAAUACAUUA- (((((((((.((....))..((((((.(((...((((.((..((....))..)).))))..))).)))))))))))))))....(((....(((....))).....)))..........- ( -30.20, z-score = -1.09, R) >droPer1.super_4 1859468 117 - 7162766 CGUGUCCACCAAAUGGCUACGCUGUAGAUGGCGGGUCAGUGUGCUCUUGGCCGCAAAUGAGCGAAUGCAUCGCGGACACUCAAAGGGUAGUUC--UGGGAAUAGACAAGGAAUAUAUAG- .((((((......(((((.((((......)))))))))(((.((.....)))))......((((.....))))))))))..........((((--(..(......)..)))))......- ( -33.10, z-score = 0.05, R) >dp4.chr3 7948364 117 - 19779522 CGUGUCCACCAAAUGGCUACGCUGUAGAUGGCGGGUCAGUGUGCUCUUGGCCGCAAAUGAGCGAAUGCAUCGCGGACACUCAAAGGGUAGUUC--UGGGAAUAGACAAGGAAUAUAUAG- .((((((......(((((.((((......)))))))))(((.((.....)))))......((((.....))))))))))..........((((--(..(......)..)))))......- ( -33.10, z-score = 0.05, R) >droAna3.scaffold_13266 6882598 108 + 19884421 CGUCUCCACCAAAUGGCUUCGCUGCAGAUGCCGAACUAAGGUGCUCUUGGCCGCAAACGAGCGAAUGCAUCGCGGGCACUCAAAGGGCAGCUC--UGGAAAUAGAUUUUA---------- ....((((.....((((.((......)).)))).....((.(((((((.((((....)..((((.....)))).))).....))))))).)).--))))...........---------- ( -29.20, z-score = 1.10, R) >droEre2.scaffold_4845 3592524 118 + 22589142 UGUAUCAACCAAAUGACUGCGCUGCAGGUGGCGACUAAGUGUGGUCUUCGCCGCAAAUGAUCGAAUGCAGCGCGGGCAUUCGAAUGGUAAUUC--UGUUGUUGGAUAUAGGAGGUUAUUC ......((((.((((.((((((((((.....(((.((..((((((....))))))..)).)))..)))))))))).))))..........(((--(((........)))))))))).... ( -40.00, z-score = -2.06, R) >droYak2.chr2R 18788710 117 + 21139217 UGUGUCCACCAAAUGACUGCGCUGCAGGUGUCGACUAAGGGUGGUCUUAGCAGCAAAUGACCGAAUGCAGCGCGGGCAUUCAAAUGGUAAUUC--UGUUGUUGGAUUUAAGAGG-UAUUA .(((.((((((((((.((((((((((....(((.((((((....))))))((.....))..))).)))))))))).))))....))))(((((--.......))))).....))-))).. ( -33.30, z-score = -0.13, R) >droSec1.super_1 4365470 117 - 14215200 UGUGUCUACCAAAUGACUGCGCUGCAGGUGGCGGCUAAGGGUGGUCUUGGCCGCAAACGAUCGAAUGCAGCGCGGGCAUUCAAAUGGUAAUUC--UGUUGUUGGACGAAAGAAG-UGUUC ......(((((((((.((((((((((..((((((((((((....)))))))))).......))..)))))))))).))))....))))).(((--(.(((.....))).)))).-..... ( -45.41, z-score = -3.02, R) >droSim1.chr2R_random 1930572 117 - 2996586 UGUGUCUACCAAAUGACUGCGCUGCAGGUGGCGGCUAAGGGUGGUCUUGGCCGCAAACGAUCGAAUGCAGCGCGGGCAUUCAAAUGGUAAUUC--UGUUGUUGGACGAAAGAAG-UGUUC ......(((((((((.((((((((((..((((((((((((....)))))))))).......))..)))))))))).))))....))))).(((--(.(((.....))).)))).-..... ( -45.41, z-score = -3.02, R) >consensus UGUGUCCACCAAAUGACUACGCUGCAGAUGGCGACUAAGGGUGCUCUUGGCCGCAAAUGAGCGAAUGCAGCGCGGACACUCAAAUGGUAAUUC__UGGAGUUGGACGAAAAA_G___UU_ .............((.((((((((((..((..((((......))))(((........))).))..)))))))))).)).......................................... (-16.44 = -15.99 + -0.45)
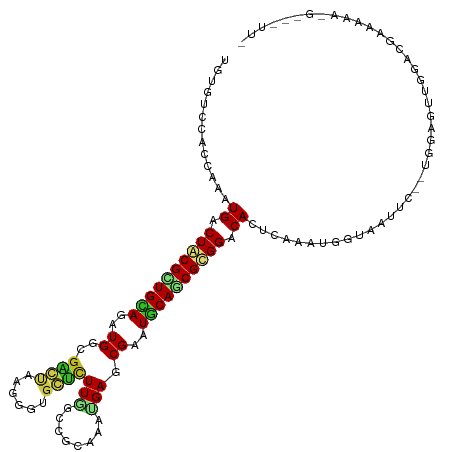
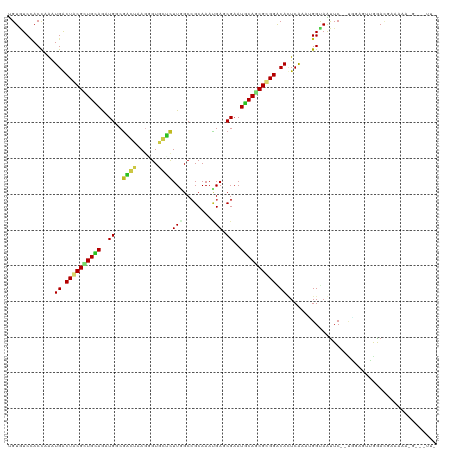
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:42 2011